8
GANs for Improving Models in Genomics
One of the significant developments in the field of Deep learning (DL) has been the introduction of new generative models. The most popular generative models are Generative Adversarial Networks (GANs), Variational Autoencoders (VAE), deep autoregressive models, style transfer, and so on. We learned about what VAEs are in the previous chapter. GANs have become a hot topic in the DL research community in the last few years. They were introduced by Ian Goodfellow in 2014 and are considered one of the most interesting ideas of the last 10 years by Yann LeCun, who is considered the father of modern DL. A GAN, as the name suggests, is a type of generative model that is trained in an adversarial setting to learn data distribution that is closer to the real world, thereby generating synthetic data inexpensively. GANs have revolutionized many domains such as natural language processing (NLP), computer vision (CV), and, most recently, genomics because of their ability to learn the data distribution and recreate artificial datasets closer to real-world data that can be used for data exploration, running tools, generating queries, test hypotheses, and so on. Since their advent, several variants of GANs were introduced, resulting in improved performance in image generation, text generation, voice synthesis, creation of artificial genomes, and so on.
Genomics data is the most complex compared to other types of data out there. Despite its complexity, the enormous potential of genomics datasets for both basic and applied research has fueled interest among genomic researchers and scientists. One of the challenge with genomics in healthcare is data privacy. GANs, because of their ability to generate artificial data derived from real-world datasets, can address data privacy. In addition, GANs can aid collaboration among researchers and the testing of ideas through open data access. GANs have several applications in genomics in addition to improving models, such as the automated design of probe sequences to perform binding assays for proteins and DNA, optimization of genomic sequences for the cell to produce a favorable chemical product, and so on. This chapter introduces GANs and how they can be used to improve models trained on genomics data. By the end of the chapter, you will know what GANs are, understand the challenges of working with genomics datasets, how GANs can improve models trained on genomics datasets, and finally, the applications of GANs in genomics.
As such, here is a list of topics that will be introduced in this chapter:
- What are GANs?
- Challenges in working with genomics datasets
- How can GANs help improve models?
- Practical applications of GANs in genomics
What are GANs?
Before we discuss GANs, you should know how generative models work. But before that, it would be advisable to understand how generative models are different from discriminative models.
Differences between Discriminative and Generative models
DL models can be broadly divided into discriminative models and generative models. Simply put, discriminative models focus on generating predictions of labels from the features mainly used for supervised learning (SL), and generative models focus on explaining how the data is generated and are used for unsupervised learning (UL). Let’s go into this a little deeper to understand the differences.
Discriminative models try to find the relationships between ![]() , such as features, and
, such as features, and ![]() , such as targets. For example, if you are trying to predict the cancer type from genomic variations (single nucleotide polymorphisms, or SNPs), the
, such as targets. For example, if you are trying to predict the cancer type from genomic variations (single nucleotide polymorphisms, or SNPs), the ![]() here indicates the features of those data instances such as the number of variations, type of variation, and so on, and the
here indicates the features of those data instances such as the number of variations, type of variation, and so on, and the ![]() here indicates cancer type. So, the discriminative model if expressed mathematically refers to
here indicates cancer type. So, the discriminative model if expressed mathematically refers to ![]() , which is the probability of a sample instance belonging to a particular cancer type
, which is the probability of a sample instance belonging to a particular cancer type ![]() based on some feature
based on some feature ![]() . As shown in Figure 8.1, during training, discriminative models learn the boundaries between classes or labels in the dataset:
. As shown in Figure 8.1, during training, discriminative models learn the boundaries between classes or labels in the dataset:

Figure 8.1 – How do discriminative models work?
The predictions from discriminative models can refer to either predicting a continuous value (regression method) or a distinct class value (classification method) as shown in the Figure 8.1. Neural network (NN) architectures such as feed-forward NNs (FNNs), convolutional NNs (CNNs), and recurrent NNs (RNNs) are examples of discriminative models.
While discriminative models learn the conditional probability ![]() , generative models learn the joint probability
, generative models learn the joint probability ![]() of the input data
of the input data ![]() and the label
and the label ![]() , and make predictions using the Bayes algorithm to calculate
, and make predictions using the Bayes algorithm to calculate ![]() . Going by the previous example, a generative model knows how the data was generated, and based on the general assumption of the data, it classifies the samples into different cancer types as shown as follows:
. Going by the previous example, a generative model knows how the data was generated, and based on the general assumption of the data, it classifies the samples into different cancer types as shown as follows:

Figure 8.2 – How do generative models work?
A popular application of a generative model in genomics is representing sequences as generative hidden Markov models (HMMs). Generative models are hard to train, and so they didn’t take off in comparison to discriminative models; however, they have played a key role in the history of machine learning (ML).
Now that you have got some background on what generative models are, let’s now dive into understanding GANs for genomics in the next few sections. But before that, let’s get some intuition about GANs first.
Intuition about GANs
GANs are a type of deep generative model that uses two competing (and cooperating) NN models referred to as a generator and a discriminator. As the name sounds, the generator generate the data distribution, and the discriminator acts as a critic and evaluates the generated data for authenticity. In other words, the discriminator decides whether the generated instance of data that it reviews belongs to the original data or not. Initially, we pass noisy random input data to the first NN (generator), which then tries to generate synthetic data that is statistically similar to the input data. On the other hand, the second NN (discriminator) is trained to differentiate between real and fake (synthetic) data (Figure 8.3):

Figure 8.3 – Intuition about GANs
As shown in the preceding diagram, the role of the generator is to generate synthetic or fake data from input data from the random noise, and the role of the discriminator, which is a fully connected NN, is to classify the generated data as real or fake.
A generator in GANs tries to generate synthetic data that resembles real data. A discriminator, on the other hand, is trained to estimate the probability distribution that the given sample comes from real data rather than the one provided by the generator (fake data or synthetic data). This way, both NNs pit against each other in an adversarial way: the generator tries to overcome the discriminator, while the discriminator tries to get better at identifying whether the generated samples are real or fake. Quite regularly, the generator tries to fool the discriminator into thinking the output data is real and asks the discriminator to label it as real data, but the discriminator naturally classifies this data as fake, as expected. The game stops when the discriminator fails to differentiate whether the output is coming from real data or the generator.
How do GANs work?
Now that you have an intuition about GANs, let’s understand how they work. The main components of GANs include the following:
The following diagram provides a visual representation of how GANs work:

Figure 8.4 – How GANs work?
Let’s get an intuition of how GANs work with the components listed in Figure 8.4:
- The generator
 transforms a latent variable
transforms a latent variable  (noisy data) to generate data
(noisy data) to generate data  . The structure of the generator
. The structure of the generator  shown in Figure 8.4 can be arbitrary. It can consist of an FNN or a CNN, or any other NN, and the only requirement is that the shape of the input matches the shape of the output data.
shown in Figure 8.4 can be arbitrary. It can consist of an FNN or a CNN, or any other NN, and the only requirement is that the shape of the input matches the shape of the output data. - Next, the discriminator
 input is either real data
input is either real data 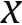 that comes from measured observations or fake data from generator
that comes from measured observations or fake data from generator  , and it outputs real number
, and it outputs real number 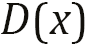 or
or  . To differentiate between the two types of data, the real data is labeled as 1.0, indicating 100% confidence in real data, and the fake data as 0.0, indicating 0% confidence in being real. The role of the discriminator is to identify real data from fake data. That means it’s a binary classifier that distinguishes between real and fake datasets. As with the generator, you can choose the NN architecture of the discriminator if the dimensions of the input and output are the same.
. To differentiate between the two types of data, the real data is labeled as 1.0, indicating 100% confidence in real data, and the fake data as 0.0, indicating 0% confidence in being real. The role of the discriminator is to identify real data from fake data. That means it’s a binary classifier that distinguishes between real and fake datasets. As with the generator, you can choose the NN architecture of the discriminator if the dimensions of the input and output are the same. - The process of training GANs is considered a zero-sum minimax game where one network’s failure is the other network’s gain. The discriminator aims to reduce the error between real and fake data, and the generator aims to increase the probability of the discriminator making a mistake. The loss of the discriminator reflects the accuracy of its predictions, and the loss of the generator is the inverse of the loss of the discriminator. The result is for the generator to produce samples closer to real-world data to fool the discriminator. Training generator and discriminator NNs at the same time is generally not stable.
Training of the discriminator
To train the discriminator NN, we label real data from the training set as 1 and fake data from the generator as 0.
The following diagram depicts the training process:

Figure 8.5 – Training of discriminator
The following is shown in Figure 8.5, for each batch of training, the following applies:
- The discriminator receives two sources of samples—real and fake data. The real data consists of images of real people, sequences of real DNA data, and so on.
- The discriminator uses these as training data. Real data is considered positive data and is provided to the discriminator during the training process. Fake data generated by the generator is treated as negative data during training.
- When the discriminator trains, the generator does not train and its parameters such as weights and biases are kept frozen. The role of the generator during that state is to provide the discriminator with the samples it needs to train.
- The discriminator is connected to two different loss functions—the discriminator’s loss and the generator’s loss. However, during discriminator training, it ignores the generator’s loss. Based on the discriminator’s loss it penalizes for wrongly classifying the real data as fake and vice versa, and the parameters are updated from the discriminator loss using backpropagation.
Training of the generator
Once the parameters of discriminator are updated so that the output is closer to the real-world data, we train the generator. The different steps in the training of the generator involve the following:
- The generator produces output from random input noise samples.
- Then, it gets feedback from the discriminator as real versus fake.
- Then, it calculates loss from the discriminator classification.
- Then, it uses backpropagation from the output to the discriminator to the generator to obtain gradients.
- Finally, it uses these gradients to adjust the parameters of the generator’s weight while keeping the discriminator’s weights unchanged.
Let’s go into the details of each of the preceding steps for the training of the generator.
You can see a visual overview of the training here:

Figure 8.6 – Training of generator
The following is shown in Figure 8.6:
- The generator produces output from random input noise samples and is then passed to the discriminator to compare it with the real output and report output loss.
- The generator gets feedback from the discriminator as to whether it is real or fake and tries to learn and improve model predictions over time.
- The generator has a loss like a discriminator, and it penalizes the generator for producing a sample that the discriminator classifies as fake. As before, the parameters of the generator are adjusted using backpropagation, and during this process, the discriminator’s parameters are kept frozen.
- When the generator does a good job to generate good data and fool the discriminator, the output probability should be close to 1.
With this background, let’s understand how GANs can help genomics but before that let's look at the current challenges working with genomic datasets.
Challenges working with genomics datasets
Genomics is the study of the genetic constitution of a whole organism, which are instructions for an organism to build and grow. It is now routinely possible to sequence a whole genome of organisms, thanks to next-generation sequencing (NGS) technologies. Despite easy access to genome sequencing technology, the primary challenge is the availability of these genomic datasets at scale because of technical limitations, cost, difficulty collecting more data, and so on. It is well known in the DL community that in general, the more data that DL can have access to, the more accurate the predictions are.
Not having enough data restricts the utility of the available data and limits building highly accurate DL models with it. Here are some of the problems arising from small data:
- Small data poses problems with model training and the use of trained models in real-world applications because it is prone to overfitting problems.
- Small data is also often confounded by problems such as class imbalance and bias, which is common in healthcare data. Class imbalance is where there is an uneven distribution of samples in one class versus another and results in models converging slower during training and poor performance on real-world datasets during model predictions.
In addition, to small data, the other big challenge for genomics remains safely sharing genomic datasets because of sensitivity and privacy issues. Our ability to safely share this data will lead to medical breakthroughs and precision medicine reality in near future.
One potential solution to address the aforementioned problems is to create synthetic datasets that can mimic real-world genomic data and can offer enhanced data privacy and enable better collaboration among genomic researchers without compromising privacy and bias. In addition, they also augment existing real-world genomic datasets. GANs can generate artificial datasets of the highly complex genomic sequences that genomic researchers routinely use. One of the main advantages of GANs is the artificial genomic datasets that have the same size and shape as the real-world dataset it was trained on, which makes GANs the preferred choice for generating synthetic data. In addition, researchers can run tools, test hypotheses, and do any other analysis on this synthetic data, similar to how they do it on real data. Before we go further, let’s understand what synthetic data is.
What is synthetic data?
As you all know, real data is obtained from direct measurement and is constrained by cost, logistics, and privacy concerns in the case of human data. In contrast, synthetic data is artificially synthesized and is closer to the real-world data generated using generative models such as GANs. Synthetic data can overcome many of the limitations of real-world data and, according to Gartner, by 2030 the use of synthetic data will overshadow real data in AI models (https://www.forbes.com/sites/robtoews/2022/06/12/synthetic-data-is-about-to-transform-artificial-intelligence/?sh=fbe727c75238). This is mainly caused by several factors such as an increase in compliance costs, regulatory restrictions because of the Health Insurance Portability and Accountability Act (HIPAA), the General Data Protection Regulation (GDPR), the California Consumer Privacy Act (CCPA), ever-increasing cybersecurity attacks, data breaches, privacy concerns, increased costs for manual annotation of data for DL. Synthetic data plays a key role in the advancement of DL, especially in genomics, because of limitations of the data shareability and a lack of relevant data, which is common in many genomic applications to train a model effectively. Synthetic data can augment real-world training data and address these limitations. There are several use cases of synthetic data, especially those instances where positive cases are very rare compared to negative cases—for example, cancer data where the number of samples from cancer is far fewer than the number of non-cancer patients.
Synthetic data for genomics
Synthetic data can augment limited genomic datasets that are available for many organisms. This way, we can improve the DL models to produce better and more accurate results and also prevent algorithm biases and overfitting. For example, it is widely known that the public datasets available out there are unbalanced in terms of gender, race, geography, and so on. Because of this imbalanced nature of datasets, the algorithm is biased toward the majority class (the class that has more data points), and classification accuracy suffers because of this. If we were to use this model, it would work great for the majority class but would have poor performance for the minority class (the class that has low data points), although its performance for the minority class is important compared to the majority class.
There are techniques such as oversampling and adding a penalty term to the cost function for the wrong prediction, which we will discuss briefly next:
- Oversampling is where instances of the minority class are duplicated. The Synthetic Minority Oversampling TEchnique (SMOTE) is a very popular oversampling technique that creates synthetic samples by interpolating between neighboring minority classes in “feature space” rather than in the “data space”. It uses the k-nearest neighbors (KNN) method and creates a synthetic sample at a random point on the line between each data point and a chosen neighbor. Even though SMOTE has been widely adopted by ML practitioners and has been successfully used for unbalanced datasets, it’s proven to be suboptimal for the generation of synthetic data for high-dimensional datasets such as NLP, CV, genomics, and so on.
- Cost-sensitive learning, where we modify the cost function to penalize the misclassification of the minority class. However, it also suffers from the same issues as oversampling method.
- Synthetic data generation using GANs. Generative models such as GANs, because of their ability to learn data distribution from the trained data, can create synthetic datasets with the same size and distribution as the original data that they have been trained on and thereby can augment high-dimensional datasets such as images, audio, videos and, genomic datasets and address class imbalance. Several published pieces of the literature showed the promise of GANs for augmenting the trained dataset and thereby improving performance in the case of imbalanced high-dimensional class-imbalance problems. The three main ways synthetic datasets can help researchers, developers, scientists, and enterprises are summarized as follows:
- Making data accessible and shareable, thereby allowing for faster and safer collaboration on the data and arriving at interesting and innovative findings sooner
- Generating more samples from the limited datasets can help models generalize well on unseen data
- Reducing bias in training datasets, which in turn can help build representative and highly accurate models thereby improving models.
You now have some background about what GANs are, how they work, and how GANs can help address some of the current challenges in genomics. With this background, let’s spend some understanding how GANs can help improve models.
How can GANs help improve models?
DL requires a lot of data to mine insights and make an informed decision. The success of DL to generalize well is mainly attributed to the training of NN architectures on large amounts of data. However, it is not always possible to acquire more data because of several reasons, as explained earlier. What if we can generate synthetic data that is modeled around real-world data so that we can augment the limited datasets and improve our model predictions? Synthetic data has a multitude of use cases in DL because of the infinite variations of synthetic data that can be produced. DL is the primary beneficiary of synthetic data, and research shows that enhancing real-world data with synthetic data produced using generative models such as GANs can significantly improve model fitness and thereby result in better predictions. GANs can help improve models directly and indirectly through the generation of synthetic data, which can make sensitive data accessible to help researchers understand the features in the data that best explain the problem. It can also be used to augment limited datasets—which are quite common in genomics because of technical limitations, costs, and feasibility—to balance data to reduce bias and improve predictions for models, which is the most important component of what GANs do.
Before we understand how GANs can help improve models, let's refresh our understanding of how synthetic data is generated. Briefly, synthetic data is generated from real-world datasets by using generative models such as GANs that look at the distribution of data points in the training data and resample data from the real-world data, as shown in Figure 8.7:

Figure 8.7 – Synthetic data generation process using GANs
Let’s see an example of how synthetic data using GANs can be produced using a real-world case study.
Scientists often explore the relationships between the phenotype (physical characteristics of an organism height, weight, and so on) and genotype (genetic differences that exist at specific locations such as SNPs and indels) to identify genes controlling a particular disease, a process termed genome-wide association studies (GWAS). One such widely-used repositories to study populations is UK Biobank, which hosts the phenotypic and genotypic data from a large number of individuals and has more than 400 billion data points. For this example, we will take data set of 1,200 mice that contains 68 phenotypes and nearly 100,000 SNPs.
Let’s now use GANs to create synthetic versions of this real-world dataset one for phenotypes and one for genotypes from the aforementioned data. Once we generate synthetic datasets, can then compare the statistical results of the real-world and synthetic datasets to find out if synthetic datasets are any good compared to real-world datasets. Here are brief the steps:
- The first step is to build a phenotype training set where we create and format real-world training data for each phenotype batch that will be generated.
- Next, from the phenotype training data, train synthetic model to generate synthetic phenotypes matching their size and shape.
- Similar to building a phenotype training set, we build a genotype training set where we format and build a training set for genomic data along with synthesized phenotype data.
- And finally, we train a synthetic model on real-world genotypic data and ensure that the synthetic phenotypes line up with our newly created synthetic genotypes.
- Once a synthetic version of the data is generated, we can do an initial analysis of this data. We can compare the accuracy of the synthetic genotype and phenotype data to the real-world training sets through a correlation matrix. We can also do principal component analysis (PCA) between the two datasets to examine how effectively the synthetic model learned the structure and distribution of the real-world data.
- Once the synthetic data is validated by comparing it with the real-world data through correlations and PCA, we can now leverage this synthetic data for genomics-related use cases.
We have just seen a use case of how GANs can help improve models by creating synthetic datasets. Similarly, there are several other applications of GANs for genomics that we will learn about in the last section of this chapter.
Practical applications of GANs in genomics
GANs have found a lot of applications in several domains such as NLP, CV, and genomics because of their ability to produce synthetic data samples to augment the real world and help improve models’ fitness. State-of-the-art synthetic models such as GANs can produce an artificial version of high-dimensional and complex genomic datasets with high accuracy, scale, and privacy. The artificial datasets can be shared among researchers and enable future genomics research and safe, private data sharing between researchers, health care providers, and the industry. As discussed briefly in the introduction, there are several use cases of GANs in genomics such as the automatic design of probe sequences for binding assays, optimization of genomic sequences, creation of synthetic genomes, and so on.
We will now see some examples of how GANs are applied to genomics and solve some real-world problems in the following section.
Analysis of ScRNA-Seq data
Single-cell RNA-Seq (scRNA-Seq) technologies have enabled gene expression profiling at a single-cell resolution level because of the advances in Single-cell and NGS technologies. ScRNA-Seq gene profiles enable the understanding of the function of genes at a single-cell resolution level like never before. There is a lot of ScRNA-Seq data that is currently available, to extract meaningful biological insights. Despite the availability of large amounts of data, integration and analysis of scRNA-Seq data is computationally challenging because of biological and technical noise coming from different laboratories and between different batches within the same experiment. Current methods such as dimensionality reduction, clustering, and so on can help find the structure in the data when the data is devoid of any noise, but they cannot help integrate the data coming from diverse laboratories and experimental protocols.
GANs can be applied to a few types of scRNA-Seq data originating from different labs and different experimental protocols and can generate realistic scRNA-Seq data that spans the full diversity of all the cell types. This is a powerful framework for the analysis of scRNA-Seq data by integrating multiple datasets irrespective of the origin and experimental protocols. Let’s understand how this is done now.
Have a look at the following diagram:

Figure 8.8 – An overview of single-cell RNA-Seq analysis using GANs
In the preceding diagram, GANs with the two NN models—the generator and discriminator—concurrently train and compete. The role of the generator, as learned before, is to transform data from the latent variable or random noise into a single-cell gene expression profile. Next, the discriminator tries to evaluate whether the generated data is closer to the real-world data or generated. Only the discriminator sees the real scRNA-Seq data, which is not corrected for batch effect and technical variation, while the generator tries to improve its synthetic data through its interaction with the discriminator. Once the GAN is optimized by adjusting the generator and discriminator parameters using backpropagation, we can extract the biological insights from the generator and discriminator NN models such as gene association networks, gene expression ranges, dimensionality reduction, and so on.
Generation of DNA
The generation of DNA is the most common application of GANs in genomics. Deep generative models such as GANs can be used to model DNA sequences. DNA sequence data is unique and can be considered a hybrid between NLP and CV. DNA is the most natural language ever, consisting of the 4 nucleotides - A, G, C, and T organized on a gene with a hierarchical structure such as introns exons. As with a CV model, they have regularly repeating patterns (motifs). GANs can be used to create DNA sequences that have desired characteristics.
The framework shown in Figure 8.9 is used to create a DNA sequence:

Figure 8.9 – Basic schematic of GAN for generation of DNA
This model consists of the same two components—generator and discriminator—and they are both used for training this architecture.
- As we have learned before, a discriminator is trained using real data and fake data. Here, the real data is taken from the real-world dataset, and the fake data has obtained from the generator by sampling the random noise data (latent variable z).
- During the training of the generator, synthetic data is generated by sampling the random noise data (latent variable z). and sent to the discriminator to be scored to maximize scores for the synthetic data.
- The discriminator tries to maximize the score of real-world data taken from the real-world dataset and minimize the scores of fake data generated by the generator.
- Both models are trained using standard gradient descent algorithms. Finally, after training, we can use the generator for the creation of synthetic data and, in this case, DNA sequence data.
Using GANs for augmenting population-scale genomics data
As we saw previously, even though NGS has made it possible to generate large-scale data through sequencing genomes quickly and cheaply, it is not always possible to generate data—for example, in the case of rare diseases where samples are limited, and so the generation of data from those samples is hard. In addition to this, data privacy is an issue when accessing human data for research and other purposes. If we decide to build models on the available data, which is small and unbalanced, then the models will be biased and conclusions will be error-prone. Previously, we saw an example of how GANs can help address this problem. Unfortunately, GANs are super hard to build in Keras, so we will not do a hands-on analysis. Readers are encouraged to check out the following link for detailed instructions on how to build GANs using Keras from a similar dataset: https://github.com/shilab/PG-cGAN.
Summary
DL algorithms have seen a major upgrade recently with the development of generative models such as VAEs and GANs, which contributed significantly to the creation of synthetic datasets. With this development, the fields of CV, NLP, and genomics have profited immensely. In the last chapter on Unsupervised learning using Autoencoders, you were introduced to VAE and in this chapter, you were introduced to GANs and how they can be used to address some of the limitations of genomics data and, improve DL models. First, we looked at the differences between discriminative and generative models, and then next we understood the key components of GANS which are the generator and discriminator, how they are trained and constantly pit against each other in an adversarial way to generate synthetic data as close as possible to real-world data.
Because of GANs ability to generate synthetic data and DL’s requirement for a large amount of data, we see how GANs are used for improving models—specifically, DL models. Then we had a case study where we understood how GANs can help create synthetic datasets to augment the phenotype and genotype real-world datasets and perform accurate GWAS analysis. Finally, we saw some of the important applications of GANs in genomics such as the analysis of heterogenous scRNA-Seq data and the creation of artificial DNA sequences. We also realized that these could not have been possible if it were not for GANs, and it’s only the start of what GANs can do for genomics.
In the next chapter, we will look at how the different deep NN (DNN) algorithms such as FNNs, CNNs, RNNs, autoencoders, and GANs that we have learned so far can be operationalized in a production environment.
