Chapter Contents
Recursive Partitioning (Decision Trees)
The Partition platform recursively partitions a data set in ways similar to carttm, chaidtm, and C4.5. Recursively partitioning data is often taught as a data mining technique, because
• it is good for exploring relationships without having a good prior model.
• it handles large problems easily.
• it works well with nominal variable and messy or unruly data.
• the results are very interpretable.
The factor columns (X’s) can be either continuous or categorical. If an X is continuous, then the splits (partitions) are created by a cutting value. The sample is divided into values below and above this cutting value. If the X is categorical, then the sample is divided into two groups of levels.
The response column (Y) can also be either continuous or categorical. If Y is continuous, then the platform fits means, and creates splits which most significantly separate the means by examining the sums of squares due to the differences in the means. If Y is categorical, then the response rates (the estimated probability for each response level) become the fitted value, and the most significant split is determined by the largest likelihood-ratio chi-square statistic (G2). In either case, the split is chosen to maximize the difference in the responses between the two branches of the split.
The Partition platform displays slightly different outputs, depending on whether the Y-variable is categorical or continuous.
Figure 17.1 shows the partition plot and tree after one split for a categorical response. Points have been colored according to their response category. Each point in the partition plot represents the response category and the partition it falls in. The x- and y-positions for each point are random within their corresponding partition. The partition width is proportional to the number of observations in the category, and the partition height is the estimated probability for that group.
Figure 17.2 shows the case for a continuous response. The points are positioned at the response value, above or below the mean of the partition they are in.
Figure 17.1 Output with Categorical Response (Titanic Passengers.jmp)

Figure 17.2 fOutput for Continuous Responses (Lipid Data.jmp)

Growing Trees
As an example of a typical analysis for a continuous response, open the Lipid Data.jmp data table. This data contains results from blood tests, physical measurements, and medical history for 95 subjects. For examples using a categorical response, see the exercises at the end.
Cholesterol tests are invasive (requiring the extraction of blood) and require laboratory procedures to obtain results. Suppose these researchers are interested in using non-invasive, external measurements and information from questionnaires to determine which patients are likely to have high cholesterol levels. Specifically, they want to predict the values stored in the Cholesterol column with information found in the Gender, Age, Height, Weight, Skinfold, Systolic BP, and Diastolic BP columns.
To begin the analysis,
Note: The default partitioning method is called the Decision Tree method. If you are using JMP Pro, a Methods menu appears at the lower left of the launch dialog, which offers two additional methods: Bootstrap Forest and Boosted Tree. JMP Pro also provides additional validation features.
Figure 17.3 Partition Dialog

Figure 17.4 shows the initial Partition report that appears. By default, the Candidates node of the report is closed, but is open here for illustration. Note that no partitioning has happened yet—all of the data are placed in a single group whose estimate is the mean cholesterol value (191.23).
Figure 17.4 Initial Lipid Partition Report

The Partition platform looks at values of each X variable to find the optimum split. The variable that results in the highest reduction in total sum of squares is the optimum split, and is used to create a new branch of the tree.
In the Candidate SS column, Diastolic BP results in a reduction of 10691.44094 in the total SS, so it is used as the splitting variable.
To begin the partitioning process, you interactively request splits.
As expected, the Diastolic BP variable splits the sample.
The complete report is in Figure 17.5. People with a diastolic blood pressure less than 80 tend to have lower cholesterol (a mean of 183.3) than those with blood pressure of 80 or more (with a mean of 205.4).
Figure 17.5 First Split of Lipid Data

An examination of the candidates in Figure 17.6 shows the possibilities for the second split. Under the Diastolic BP<80 leaf, a split using the Weight variable has a LogWorth of 1.76. The highest LogWorth under the Diastolic BP >=80 leaf is 1.21 for the Age variable. Therefore you expect that pressing the split button again will give two new weight leaves under the Diastolic<80 leaf, since Weight has the highest overall LogWorth.
Figure 17.6 Candidates for Second Split

The resulting report is shown in Figure 17.7, with its corresponding tree shown in Figure 17.8.
Figure 17.7 Plot After Second Split

Figure 17.8 Tree After Second Split

This second split shows that of the people with diastolic blood pressure less than 80, weight is the best predictor of cholesterol. For this group, the model predicts that those who weigh 185 pounds or more have an average cholesterol of 149. For those that weigh less than 185 pounds, the predicted average cholesterol is 188.5.
You can continue to split until you are satisfied with the predictive power of the model. As opposed to software that continues splitting until a criterion is met, JMP allows you to be the judge of the effectiveness of the model.
Viewing Large Trees
With several levels of partitioning, tree reports can become quite large. JMP has several ways to ease the viewing of these large trees.
• Use the scroll tool ( ) found on the tool bar or Tools menu to easily scroll around the report.
) found on the tool bar or Tools menu to easily scroll around the report.
• Use the Display Options from the Partition red triangle menu to turn off parts of the report that are not needed. As an example, Figure 17.9 shows the current lipid results after four splits, with Display Options > Show Split Stats and Display Options > Show Split Candidates turned off.
Figure 17.9 Lipid Data After Four Splits

You can also request a more compact version of the partition tree.
This option toggles a compact view of the tree, appended to the right of the main partition graph. Figure 17.10 shows the Small Tree View corresponding to Figure 17.9.
Figure 17.10 Small Tree View

Note: The font used for the small tree view is controlled by the Small font, found on the Fonts tab of the JMP preferences. Figure 17.10 uses an 8-point Arial font. Control-Shift + (or Command + on the Macintosh) can also be used to increase font sizes.
Viewing Column Contributions
You might want to see a summary of how many times a variable was split, along with the sums of squares attributed to that variable. This is particularly useful when building large trees involving many variables. The Column Contribution report provides this information. To see the report in Figure 17.11 choose Column Contributions from the top red triangle menu.
Figure 17.11 Column Contributions After Four Splits

Exploratory Modeling with Partition
You might wonder if there is an optimum number of splits to do for a given set of data. One way to explore recursive splitting is to use K Fold Crossvalidation. Lets do that now.
• check Split History
• uncheck Display Options > Show Tree
• uncheck Display Options > Show Graph
• check K Fold Crossvalidation. When the cross validation window shows, use the default of 5 subgroups and click OK.

You have now tailored the Partition platform to let you interactively split or prune the model step-by-step and observe the results. Furthermore, you are using a K Fold validation that randomly divides the original data into K subsets. Each of the K subsets is used to validate the model fit on the rest of the data, fitting a total of K models. The model giving the best validation R2 statistic is considered the best model. The beginning partition platform should look like Figure 17.12.
Figure 17.12 Initial Partition Platform with Cross Validation and History Plot

Figure 17.13 shows the Split History after eight splits for this example. Your validation results might not be exactly the same as those shown because the validation subsets are randomly chosen.
If you continue to split until the data is completely partitioned, the model continues to improve fitting the data. However, this usually results in overfitting, which means the model predicts the fitted data well, but predicts future observations poorly, as seen by less desirable validation statistics.
We can see this in our example (Figure 17.13). After 8 splits, the Overall R2 is 0.455, but the K Fold R2 is 0.27. From split 7 to split 8 the Overall R2 increased, but the validation R2 decreased. This could be an example of overfitting.
Figure 17.13 Partition platform with Validation after Eight Splits

Saving Columns and Formulas
Once you have partitioned the data to your satisfaction, you can save the partition model information.
The Save Columns submenu shows the options for saving results. All commands create a new column in the data table for storing their values. The commands that contain the word Formula (Save Prediction Formula, for example) store a formula in the column, where the other commands save values only. As an example,

This adds a column to the report named Cholesterol Predictor that contains a formula using the estimates from the partitions of the tree.
To see the formula, return to the Lipid Data table and
Figure 17.14 Formula for Partition Model Example After Four Splits

You can see that the formula is a collection of nested If functions that use the partitioning results. You can copy and paste the formula to test its validity on other similar data.
Neural Nets
The Neural platform implements a connected network with one layer (or two layers in JMP Pro). Neural networks can predict one or more response variables using flexible functions of the input variables. This kind of model can be a very useful when it is not necessary to
• describe the functional form of the response surface,
• to describe the relationship between the input variables and the response, or
• narrow down the list of important variables. 

A neural network can be thought of as a function of a set of derived inputs called hidden nodes. Hidden nodes are nonlinear functions (called activation functions) of the original inputs. You can specify as many nodes as you want.
At each node, the activation function transforms a linear combination of all the input variables using the S-shaped hyperbolic tangent function (TanH). The function then applied to the response is a linear combination of the nodes for continuous responses or a logistic transformation for categorical responses.
Note: Additional activation functions and other features are available in JMP Pro. This section continues with a simple example showing the abilities of the Neural platform in JMP. For more advanced examples that use JMP Pro features and technical details of the particular functions used in the neural network implementation, see the JMP Modeling and Multivariate Methods book (use Help > Books).
A Simple Example
This section uses the Peanuts.jmp sample data, from an experiment that tests a device for shelling peanuts. A reciprocating grid automatically shells the peanuts. The length and frequency of the reciprocating stroke, as well as the spacing of the peanuts, are factors in the experiment. Kernel damage, shelling time, and the number of unshelled peanuts need to be predicted. This example illustrates the procedure using only the number of unshelled peanuts as the response. A more involved neural modeling situation could have several factors, multiple responses, or both..
Note: If you are using JMP Pro, or earlier versions of the Neural platform, your initial Control Panel has different options.
When the Control Panel appears, you have the option of selecting the validation method best suited for your data.
The options on the Validation Method menu (middle of Figure 17.15) determine the way the Neural fitting machinery subsets your data to test and decide on a final model:
• Excluded Rows Holdback uses row states to subset the data. Unexcluded rows are used as the training set, and excluded rows are used as the validation set.
• Holdback, the default, randomly divides the original data into the training and validation sets. You can specify the proportion of the original data to use as the validation set (holdback). The holdback proportion 0.333 is the default.
• KFold divides the original data into K subsets. Each of the K sets is used to validate the model fit on the rest of the data, fitting a total of K models. The model giving the best validation statistic is chosen as the final model.
Holdback, the default option, is the best used for larger samples (hundreds or thousands of observations). The KFold option is better used for smaller samples such as the peanuts data.
Figure 17.15 Neural Control Panel with Validation Options

Your results will be different because the Neural fitting process begins with a random seed. The random seed determines the starting point for the search algorithm.
Figure 17.16 Example Results from Neural Platform

The reports in Figure 17.16 give straightforward information for both the training and validation samples. You can see that the KFold number is reflected by 16 subjects in the training group and 4 in the validation group. The RSquare for the training group of 0.836 is very good, and the high RSquare for the validation group of 0.854 gives confidence that the model fits well. A Validation RSquare that is substantially lower than the Training RSquare indicates that the model is fitting noise rather than structure.
Neural networks are very flexible functions that have a tendency to overfit the data. When that happens the model predicts the fitted data well, but predicts future observations poorly. However, the Neural platform has a penalty system built in the helps prevent the consequences of over fitting. If you are running JMP Pro, there is an option to pick a specific penalty function.
You can use Model Launch, shown open in Figure 17.16, and click Go to run the neural net fitting as many times as you want, or to run models with different numbers of nodes.
You can also request a diagram of the example showing how the factor columns are transformed through three hidden nodes, whose outputs are then combined to form the predicted values.
You should see the diagram shown in Figure 17.17.
Figure 17.17 Neural Net Diagram
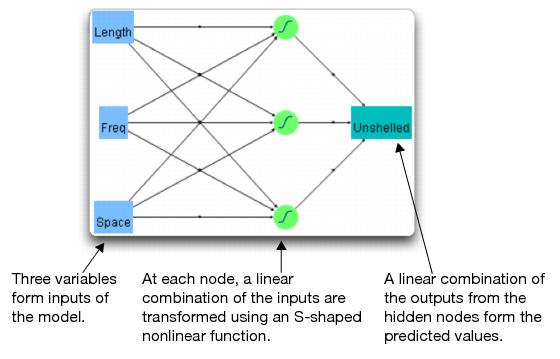
Modeling with Neural Networks
In the Neural Net diagram its easy to see the inputs and the output, but the circle nodes might look more like black boxes. However, red triangle menu options for the Model let you see what is in the nodes and how they are used to produce the predicted output.
Saving Columns
Like most analysis platforms, results from the Neural analyses can be saved to columns in the data table. The following save options are available.
Save Formulas creates new columns in the data table that contain formulas for the predicted response and for the hidden layer nodes. This option is useful if rows are added to the data table, because predicted values are automatically calculated.
Save Profile Formulas creates new columns in the data table that contain formulas for the predicted response. Formulas for the hidden nodes are embedded in this formula. This option produces formulas that can be used by the Flash version of the Profiler.
Save Fast Formulas creates a new columns in the data table for the response variable, with embedded formulas for the hidden nodes. This formula evaluates faster that the Save Profile Formula results but cannot be used in the Flash version of the Profiler.
For this example, the new column, called Predicted Unshelled, contains the formula shown in Figure 17.18. You can see three TanH formulas for the three hidden nodes embedded in this prediction formula.
Figure 17.18 Saved Profile Formula

The Actual by Predicted and the Residual by Predicted plots are similar to their counterparts in linear regression. Use them to help judge the predictive ability of the model. In this example, the model fits fairly well, and there are no glaring problems in the residual plots.
Figure 17.19 Neural Net Plots

Profiles in Neural
The slices through the response surface can be interesting and informative.
The Prediction Profiler (Figure 17.20) clearly shows the nonlinear nature of the model. Running the model with more hidden nodes increases the flexibility of these curves; running with fewer stiffens them.
The Profiler has all of the features used in analyzing linear models and response surfaces (discussed in Analyze the Model of this book and in the JMP Modeling and Multivariate Methods book under Help > Books).
Since we are interested in minimizing the number of unshelled peanuts, we use the Profiler’s Desirability Functions, which are automatically shown (Figure 17.20).
Figure 17.20 Prediction Profiler with Initial Settings

To have JMP automatically compute the optimum factor settings,
JMP computes the maximum desirability, which in this example is a low value of Unshelled. Results are shown in Figure 17.21.
Figure 17.21 Prediction Profiler with Maximized Desirability Settings

Optimal settings for the factors show in red below the plots. In this case, optimal Unshelled values came from setting Length =2.5, Freq = 127.87, and Space = 0.48. The predicted value of the response, Unshelled, dropped from 175.14 to 42.51.
Note: Recall that your results will be different from the example shown here because a random seed is used to determine the starting point for the Neural fitting process.
In addition to seeing two-dimensional slices through the response surface, the Contour Profiler can be used to visualize contours (sometimes called level curves) and mesh plots of the response surface. The Surface Profilers also gives an interesting vies of the response surface.
There are options on the Model red triangle menu for both of these plots. However, if you saved the prediction formula for the Neural model fit, you can replicate all the model plots at any time using commands in the Graph menu. Earlier, we showed how to save the profile formula in a single column (see Figure 17.18). You should have a column in the Peanuts table called Predicted Unshelled. Lets use Graph commands to see more views of the results.
You will see results that are similar to those in Figure 17.22. If necessary, use the grabber (hand tool) and drag to adjust the axes so that you can see the range of specified values.
You can see that for these low and high limits and factor settings, the number of unshelled peanuts falls in the acceptable (unshaded) region. Click on the cross-hairs and drag to explore the region for the two variables selected.
Figure 17.22 Contour Profiler with Optimal Values

You can also see a response surface view of the number of unshelled peanuts as a function of two of the three factors, using a Surface Profiler or Surface Plot (which has more features). The Surface Profiler is available in the Neural platform, or from any of the profilers available from the Graph menu. The Surface Plot command on the Graph menu can be used if you saved the prediction equation, as in this example.
The surface plot in Figure 17.23 shows the surface when the optimal settings for the factors and the predicted value of unshelled peanuts are entered into the platform, as shown.
This plot shows just how nonlinear, or bendy, our fitted Neural model is.
Figure 17.23 Surface Plot of Unshelled Peanuts

Exercises
1. As shown in this chapter, the Lipid Data.jmp sample data set contains blood measurements, physical measurements, and questionnaire data from subjects in a California hospital. Repeat the Partition analysis of this chapter to explore models for
(a) HDL (good cholesterol - higher than 60 is considered protection against heart disease)
(b) LDL (bad cholesterol - less than 120 is optimal)
(c) Triglyceride levels (less than 150 is normal)
Statistics provided by the American Heart Association (www.heart.org).
2. Use the Peanuts.jmp sample data set and the Neural platform to complete this exercise. The factors of Freq, Space, and Length are described earlier in this chapter. Use these factors for the following:
(a) Create a model for Time, the time to complete the shelling process. Use the Profiler and Desirability Functions to find the optimum settings for the factors that minimize the time of shelling.
(b) Create a model for Damaged, the number of damaged peanuts after shelling is complete. Find values of the factors that minimize the number of damaged peanuts.
(c) Compare the values found in the text of the chapter (Figure 17.21) and the values you found in parts (a) and (b) of this question. What settings would you recommend to the manufacturer?
3. The file Mushroom.jmp contains 22 characteristics of 8,124 edible and poisonous mushroom. To see probabilities, select Display Options > Show Split Prob from the top red triangle menu.
(a) Use the Partition platform to build a 7 split model to predict whether or not a mushroom will be edible based on the 22 characteristics.
(b) Which characteristics are most important? Hint: Use Column Contributions.
(c) What are the characteristics of edible mushrooms? Hint: Use the Small Tree View and Leaf Report.
(d) Prune back to 2 splits. What is the predicted probability that a mushroom with no odor and a white sport print color will be edible?
(e) Use K Fold Crossvalidation to determine the best number of splits. Do the validation statistics improve much after 3 splits? After 4 splits?
Exploratory Modeling: A case study
The following exercise is a case study that examines characteristics of passengers on the Titanic. The response is whether an individual passenger survived or was lost. The case study uses many of the platforms introduced so far in this book.
4. Open the Titanic Passengers.jmp file (use Help > Sample Data), which describes the survival status of individual passengers on the Titanic. The response variable is Survived (“Yes” or “No”), and the variables of interest (factors or x-variables) are Passenger Class, Sex, Age, Siblings and Spouses, and Parents and Children.
(a) Use the Distribution platform and dynamic linking to explore the Survived variable and the variables listed above. Click on the bars for “Yes” and “No” in the Survived plot. Does Survived seem to be related to any of the other variables?
(b) Use the Fit Y by X and Graph Builder platforms to further explore the relationship between Survived and the other variables.
• Did passengers who survived tend to fall in a particular Passenger Class?
• Did they tend to be males or females?
• Is Survived related to the other variables?
• Do there appear to be any interactions? For example, does the relationship between Passenger Class and Survived depend on the Sex?
(c) Use Fit Model to fit a logistic model for the response Survived and the five factors. Include the following interaction terms: Passenger Class*Sex, Passenger Class*Age, Sex*Age.
• Are any of the interactions significant?
• Which main effects are significant?
• Use the profiler (under the top red triangle menu) to explore the model. Drag the vertical red lines for each factor to see changes in the predicted survival rate. Since interactions were included, you will also see changes in the profile traces for other factors.
• For which group was the predicted survival rate the highest?
• For which group was it the lowest? (Keep this window open).
(d) Use the Partition platform to build a classification tree for Survived, using the same five factors.
• Select Display Options > Show Split Prob from the top red triangle menu, then split the model several times.
• What are the most important split variables?
• Do you see evidence of important interactions? For example, were the second and third splits on the same variable, or did it choose different split variables?
• Compare these results to those found earlier using logistic regression. Do you come to similar conclusion, or are the conclusions very different? (Keep this window open as well).
(e) Use the Neural platform to build a Neural Net for Survived, using the same factors. Click Go on the Neural control panel to accept the default model settings.
• Select the Profiler from the Model red triangle menu. Drag the vertical red lines for each factor to explore changes in the predicted survival rate.
• How does this profiler compare to the one for the Logistic model found previously?
(f) Using the three final models (logistic, partition, and neural), to determine the predicted survival rate for (1) a first class female and (2) a 20 year old man. Are the results comparable?
(g) Summarize your exploration of the Titanic data and conclusions in a form suitable for presentation. (Note: In JMP, a Journal is can be an effective way to save and share results. To add content to a journal, select File > New > Journal. Then, in the Journal window right-click and select Add All Open Files.)
..................Content has been hidden....................
You can't read the all page of ebook, please click here login for view all page.
