Up to this point, we have seen how to cluster genetic variants. We have also used the Elbow method and found the number of optimal k, the tentative number cluster. Now we should explore another task that we planned at the beginning—that is, ethnicity prediction.
In the previous K-means section, we prepared a Spark DataFrame named schemaDF. That one cannot be used with H2O. However, an additional conversion is necessary. We use the asH2OFrame() method to convert the Spark DataFrame into an H2O frame:
val dataFrame = h2oContext.asH2OFrame(schemaDF)
Now, one important thing you should remember while using H2O is that if you do not convert the label column into categorical, it will treat the classification task as regression. To get rid of this, we can use the toCategoricalVec() method from H2O. Since H2O frames are resilient, we can further update the same frame:
dataFrame.replace(dataFrame.find("Region"),
dataFrame.vec("Region").toCategoricalVec()).remove()
dataFrame.update()
Now our H2O frame is ready to train an H2O-based DL model (which is DNN, or to be more specific, a deep MLP). However, before we start the training, let's randomly split the DataFrame into 60% training, 20% test, and 20% validation data using the H2O built-in FrameSplitter() method:
val frameSplitter = new FrameSplitter(
dataFrame, Array(.8, .1), Array("training", "test", "validation")
.map(Key.make[Frame]),null)
water.H2O.submitTask(frameSplitter)
val splits = frameSplitter.getResult
val training = splits(0)
val test = splits(1)
val validation = splits(2)
Fantastic! Our train, test, and validation sets are ready, so let us set the parameters for our DL model:
// Set the parameters for our deep learning model.
val deepLearningParameters = new DeepLearningParameters()
deepLearningParameters._train = training
deepLearningParameters._valid = validation
deepLearningParameters._response_column = "Region"
deepLearningParameters._epochs = 200
deepLearningParameters._l1 = 0.01
deepLearningParameters._seed = 1234567
deepLearningParameters._activation = Activation.RectifierWithDropout
deepLearningParameters._hidden = Array[Int](128, 256, 512)
In the preceding setting, we have specified an MLP having three hidden layers with 128, 256 and 512 neurons respectively. So altogether, there are five layers including the input and the output layer. The training will iterate up to 200 epoch. Since we have used too many neurons in the hidden layer, we should use the dropout to avoid overfitting. To avoid achieve a better regularization, we used the l1 regularization.
The preceding setting also states that we will train the model using the training set, and additionally the validation set will be used to validate the training. Finally, the response column is Region. On the other hand, the seed is used to ensure reproducibility.
So all set! Now let's train the DL model:
val deepLearning = new DeepLearning(deepLearningParameters)
val deepLearningTrained = deepLearning.trainModel
val trainedModel = deepLearningTrained.get
Depending on your hardware configuration, it might take a while. Therefore, it is time to rest and get some coffee maybe! Once we have the trained model, we can see the training error:
val error = trainedModel.classification_error()
println("Training Error: " + error)
>>>
Training Error: 0.5238095238095238
Unfortunately, the training was not that great! Nevertheless, we should try with different combination of hyperparameters. The error turns out to be high though, but let us not worry too much and evaluate the model, compute some model metrics, and evaluate model quality:
val trainMetrics = ModelMetricsSupport.modelMetrics[ModelMetricsMultinomial](trainedModel, test)
val met = trainMetrics.cm()
println("Accuracy: "+ met.accuracy())
println("MSE: "+ trainMetrics.mse)
println("RMSE: "+ trainMetrics.rmse)
println("R2: " + trainMetrics.r2)
>>>
Accuracy: 0.42105263157894735
MSE: 0.49369297490740655
RMSE: 0.7026328877211816
R2: 0.6091597281983032
Not so high accuracy! However, you should try with other VCF files and by tuning the hyperparameters too. For example, after reducing the neurons in the hidden layers and with l2 regularization and 100 epochs, I had about 20% improvement:
val deepLearningParameters = new DeepLearningParameters()
deepLearningParameters._train = training
deepLearningParameters._valid = validation
deepLearningParameters._response_column = "Region"
deepLearningParameters._epochs = 100
deepLearningParameters._l2 = 0.01
deepLearningParameters._seed = 1234567
deepLearningParameters._activation = Activation.RectifierWithDropout
deepLearningParameters._hidden = Array[Int](32, 64, 128)
>>>
Training Error: 0.47619047619047616
Accuracy: 0.5263157894736843
MSE: 0.39112548936806274
RMSE: 0.6254002633258662
R2: 0.690358987583617
Another improvement clue is here. Apart from these hyperparameters, another advantage of using H2O-based DL algorithms is that we can take the relative variable/feature importance. In previous chapters, we have seen that when using a Random Forest algorithm in Spark, it is also possible to compute the variable importance.
Therefore, the idea is that if your model does not perform well, it would be worth dropping less important features and doing the training again. Now, it is possible to find the feature importance during supervised training. I have observed this feature importance:
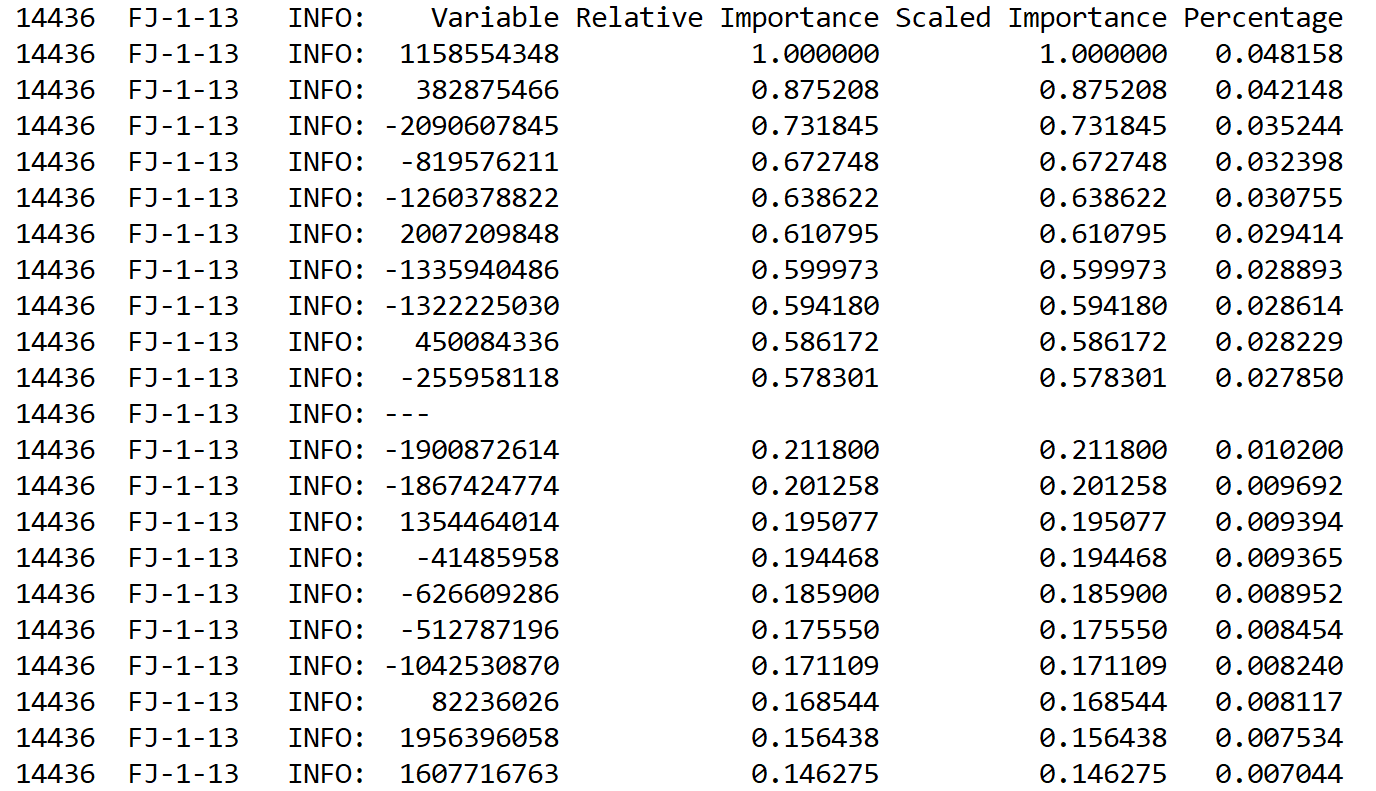
Now the question would be why don't you drop them and try training again and observe if the accuracy has increased or not? Well, I leave it up to the readers.
