Overview of Principal Component Analysis
A principal component analysis models the variation in a set of variables in terms of a smaller number of independent linear combinations (principal components) of those variables.
If you want to see the arrangement of points across many correlated variables, you can use principal component analysis to show the most prominent directions of the high-dimensional data. Using principal component analysis reduces the dimensionality of a set of data. Principal components representation is important in visualizing multivariate data by reducing it to graphable dimensions. Principal components is a way to picture the structure of the data as completely as possible by using as few variables as possible.
For p variables, p principal components are formed as follows:
• The first principal component is the linear combination of the standardized original variables that has the greatest possible variance.
• Each subsequent principal component is the linear combination of the variables that has the greatest possible variance and is uncorrelated with all previously defined components.
Each principal component is calculated by taking a linear combination of an eigenvector of the correlation matrix (or covariance matrix or sum of squares and cross products matrix) with the variables. The eigenvalues represent the variance of each component.
The Principal Components platform enables you to conduct your analysis on the correlation matrix, the covariance matrix, or the unscaled data. You can also conduct Factor Analysis within the Principal Components platform. See the Factor Analysis chapter in the Consumer Research book for details.
Example of Principal Component Analysis
To view an example Principal Component Analysis report for a data table for two factors:
1. Select Help > Sample Data Library and open Solubility.jmp.
2. Select Analyze > Multivariate Methods > Principal Components.
The Principal Components launch window appears.
3. Select all of the continuous columns and click Y, Columns.
4. Keep the default Estimation Method.
5. Click OK.
The Principal Components on Correlations report appears.
Figure 4.2 Principal Components on Correlations Report

The report gives the eigenvalues and a bar chart of the percent of the variation accounted for by each principal component. There is a Score Plot and a Loadings Plot as well. See “Principal Components Report” for details.
Launch the Principal Components Platform
Launch the Principal Components platform by selecting Analyze > Multivariate Methods > Principal Components. Principal Component analysis is also available using the Multivariate and the Scatterplot 3D platforms.
The example described in “Example of Principal Component Analysis” uses all of the continuous variables from the Solubility.jmp sample data table.
Figure 4.3 Principal Components Launch Window

Y, Columns
Lists the variables to analyze for components.
Z, Supplementary Variable
Lists the supplementary variables to be displayed. Supplementary variables are not included in the calculation of principal components and including them does not affect the results. The supplementary variables can be projected on to the loading plot and used to enhance interpretation.
Weight and Freq
Enables you to weight the analysis to account for pre-summarized data.
Note: The Weight and Freq roles are ignored for the Wide and Sparse estimation methods.
By
Creates a Principal Component report for each value specified by the By column so that you can perform separate analyses for each group.
Estimation Method
Lists different methods for calculating the correlations. Several of these methods address the treatment of missing data. See “Estimation Methods”.
Number of Components
(Appears only when Sparse is the Estimation Method.) The number of components to be estimated. See “Sparse”
Estimation Methods
Use the estimation method that addresses your specific needs. Methods are available to handle missing values, outliers, wide data, and sparse data.
You can also estimate missing values in the following ways:
• Use the Impute Missing Data option found under Multivariate Methods > Multivariate. See “Impute Missing Data” in the “Correlations and Multivariate Techniques” chapter.
• Use the Multivariate Normal Imputation or Multivariate SVD Imputation utilities found in Analyze > Screening > Explore Missing Values. See the Modeling Utilities chapter in the Predictive and Specialized Modeling book for details.
Default
The Default option uses either the Row-wise, Pairwise, or REML methods:
• Row-wise is used for data tables with no missing values.
• Pairwise is used in the following circumstances:
‒ the data table has more than 10 columns or more than 5,000 rows and has missing values
‒ the data table has more columns than rows and has missing values
• REML is used otherwise.
REML
REML (restricted maximum likelihood) estimates are less biased than the ML (maximum likelihood) estimation method. The REML method maximizes marginal likelihoods based on error contrasts. The REML method is often used for estimating variances and covariances. The REML method in the Principal Components platform is the same as the REML estimation of mixed models for repeated measures data with an unstructured covariance matrix. See the documentation for SAS PROC MIXED about REML estimation of mixed models.
REML uses all of your data, even if missing values are present, and is most useful for smaller datasets. Because of the bias-correction factor, this method is slow if your dataset is large and there are many missing data values. If there are no missing cells in the data, then the REML estimate is equivalent to the sample covariance matrix.
Note: If you select REML and your data table contains more columns than rows, JMP switches the Estimation Method. If there are no missing values, the Estimation Method switches to Row-wise. If there are missing values, then the Estimation Method switches to Pairwise.
ML
The maximum likelihood estimation method (ML) is useful for large data tables with missing cells. The ML estimates are similar to the REML estimates, but the ML estimates are generated faster. Observations with missing values are not excluded. For small data tables, REML is preferred over ML because REML’s variance and covariance estimates are less biased.
Note: If you select ML and your data table contains more columns than rows, JMP switches the Estimation Method. If there are no missing values, the Estimation Method switches to Row-wise. If there are missing values, then the Estimation Method switches to Pairwise.
Robust
Robust estimation is useful for data tables that might have outliers. For statistical details, see “Robust”.
Note: If you select Robust and your data table contains more columns than rows, JMP switches the Estimation Method. If there are no missing values, the Estimation Method switches to Row-wise. If there are missing values, then the Estimation Method switches to Pairwise.
Row-wise
Row-wise estimation does not use observations containing missing cells. This method is useful in the following situations:
• Checking compatibility with JMP versions earlier than JMP 8. Row-wise estimation was the only estimation method available prior to JMP 8.
• Excluding any observations that have missing data.
Pairwise
Pairwise estimation performs correlations for all rows for each pair of columns with nonmissing values.
Wide
Note: The Wide method extracts components based on the standardized data. The On Covariance and On Unscaled options are not available.
The Wide method is useful when you have a very large number of columns in your data. It uses a computationally efficient algorithm that avoids calculating the covariance matrix. The algorithm is based on the singular value decomposition. For additional background, see “Wide Linear Methods and the Singular Value Decomposition” in the “Statistical Details” appendix.
Consider the following notation:
• n = number of rows
• p = number of variables
• X = n by p matrix of data values
The number of nonzero eigenvalues, and consequently the number of principal components, equals the rank of the correlation matrix of X. The number of nonzero eigenvalues cannot exceed the smaller of n and p.
When you select the Wide method, the data are standardized. To standardize a value, subtract its mean and divide by its standard deviation. Denote the n by p matrix of standardized data values by Xs. Then the covariance matrix of the standardized data is the correlation matrix of X and it is given as follows:
Using the singular value decomposition, Xs is written as UDiag(Λ)V’. This representation is used to obtain the eigenvectors and eigenvalues of Xs’Xs. The principal components, or scores, are given by  .
.
Note: If there are missing values and you select the Wide method, then the rows that contain missing values are deleted and the Wide method is applied to the remaining rows.
Note: When you select the Default estimation method and enter more than 500 variables as Y, Columns, a JMP Alert recommends that you switch to the Wide estimation method. This is because computation time can be considerable when you use the other methods with a large number of columns. Click Wide to switch to the Wide method. Click Continue to use the method you originally selected.
Note: The Sparse method extracts components based only on the standardized data. The On Covariance and On Unscaled options are not available.
The Sparse method is useful when your data are sparse, meaning that they contain many zeros. It can also reduce computational time when there are a large number of columns in the data. Similar to the Wide method, the Sparse method is based on singular value decomposition. Therefore, the algorithm for the Sparse method avoids computing the covariance matrix and is computationally efficient.
Consider the same notation and standardization of X that is described in “Wide”. The correlation matrix of X is represented by the covariance matrix of Xs:
The Sparse method differs from the Wide method in the calculation of the singular value decomposition. The Wide method performs a full singular value decomposition. However, the Sparse method uses an algorithm that computes only the first specified number of singular values and singular vectors in the singular value decomposition. Therefore, only the first specified number of eigenvalues and principal components are returned. For more information about the algorithm, see Baglama and Reichel (2005).
When you select Sparse as the estimation method in the launch window, the Number of Components option appears. By default, the Number of Components is 2. Typically, the Number of Components is much smaller than the dimension of your data.
Principal Components Report
If you selected any estimation method other than Wide or Sparse, the Principal Components: on Correlations report initially appears. (The title of this report changes if you select on Covariances or on Unscaled for the Principal Components option in the Principal Components red triangle menu.)
If you select the Wide method, the Wide Principal Components report appears. If you select the Sparse method, the Sparse Principal Components report appears.
The initial Principal Components report is for an analysis on Correlations. It summarizes the variation of the specified Y variables with principal components. See Figure 4.4. You can switch to an analysis based on the covariance matrix or unscaled data by selecting the Principal Components option from the red triangle menu.
Based on your selection, the principal components are derived from an eigenvalue decomposition of one of the following:
• the correlation matrix
• the covariance matrix
• the sum of squares and cross products matrix for the unscaled and uncentered data
The details in the report show how the principal components absorb the variation in the data. The principal component points are derived from the eigenvector linear combination of the variables.
Figure 4.4 Principal Components on Correlations Report

The report gives the eigenvalues and a bar chart of the percent of the variation accounted for by each principal component. There is a Score Plot and a Loadings Plot as well. The eigenvalues indicate the total number of components extracted based on the amount of variance contributed by each component.
The Score Plot graphs each component’s calculated values in relation to the other, adjusting each value for the mean and standard deviation.
The Loadings Plot graphs the unrotated loading matrix between the variables and the components. The closer the value is to 1 the greater the effect of the component on the variable.
By default, the report shows the Score Plot and the Loadings Plot for the first two principal components. Use the list next to Select component to specify the principal components that are graphed on the Score Plot and the Loadings Plot.
Principal Components Report Options
The Principal Components red triangle menu contains the following options:
Note: Some of the options are not available for the Wide or Sparse estimation methods.
Principal Components
(Not available for the Wide or Sparse estimation methods.) Enables you to create the principal components based on Correlations, Covariances, or Unscaled.
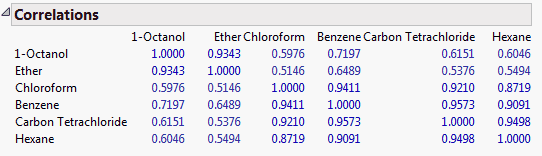
Correlations
(Not available for the Wide or Sparse estimation methods.) The matrix of correlations between the variables.
Note: The values on the diagonals are 1.0.
Figure 4.5 Correlations
Covariance Matrix
(Not available for the Wide or Sparse estimation methods.) Shows or hides the covariances of the variables.
Figure 4.6 Covariance Matrix

Eigenvalues
Lists the eigenvalue that corresponds to each principal component in order from largest to smallest. The eigenvalues represent a partition of the total variation in the multivariate sample.
The scaling of the eigenvalues depends on which matrix you select for extraction of principal components:
‒ For the on Correlations option, the eigenvalues are scaled to sum to the number of variables.
‒ For the on Covariances options, the eigenvalues are not scaled.
‒ For the on Unscaled option, the eigenvalues are divided by the total number of observations.
If you select the Bartlett Test option from the red triangle menu, hypothesis tests (Figure 4.9) are given for each eigenvalue (Jackson, 2003).
Figure 4.7 Eigenvalues

Eigenvectors
Shows or hides a table of the eigenvectors for each of the principal components, in order, from left to right. Using these coefficients to form a linear combination of the original variables produces the principal component variables. Following the standard convention, eigenvectors have norm 1.
Note: The number of eigenvectors shown is equal to the rank of the correlation matrix, or, if the Sparse method is selected, the number of components specified on the launch window.
Figure 4.8 Eigenvectors

Bartlett Test
(Not available for the Wide or Sparse estimation methods.) Shows or hides the results of the homogeneity test (appended to the Eigenvalues table). The test determines whether the eigenvalues have the same variance by calculating the Chi-square, degrees of freedom (DF), and the p-value (prob > ChiSq) for the test. See Bartlett (1937, 1954).
Figure 4.9 Bartlett Test

Loading Matrix
Shows or hides a table of the loadings for each component. These values are graphed in the Loading Plot.
The scaling of the loadings depends on which matrix you select for extraction of principal components:
‒ For the on Correlations option, the ith column of loadings is the ith eigenvector multiplied by the square root of the ith eigenvalue. The i,jth loading is the correlation between the ith variable and the jth principal component.
‒ For the on Covariances option, the jth entry in the ith column of loadings is the ith eigenvector multiplied by the square root of the ith eigenvalue and divided by the standard deviation of the jth variable. The i,jth loading is the correlation between the ith variable and the jth principal component.
‒ For the on Unscaled option, the jth entry in the ith column of loadings is the ith eigenvector multiplied by the square root of the ith eigenvalue and divided by the standard error of the jth variable. The standard error of the jth variable is the jth diagonal entry of the sum of squares and cross products matrix divided by the number of rows (X’X/n).
Note: When you are analyzing the unscaled data, the i,jth loading is not the correlation between the ith variable and the jth principal component.
Figure 4.10 Loading Matrix

Note: The degree of transparency for the table values indicates the distance of the absolute loading value from zero. Absolute loading values that are closer to zero are more transparent than absolute loading values that are farther from zero.
Formatted Loading Matrix
Shows or hides a table of the loadings for each component. The table is sorted in order of decreasing loadings on the first principal component. Therefore, the variables are listed in the order of decreasing loadings on the first component.
Figure 4.11 Formatted Loading Matrix

Tip: Use the sliders to dim loadings whose absolute values fall below your selected value and to set the degree of transparency for the loadings.
Squared Cosines of Variables
Shows or hides a table that contains the squared cosines of variables. The sum of the squared cosine values across principal components is equal to one for each variable. The squared cosines enable you to see how well the variables are represented by the principal components. You can also determine how many principal components are necessary to represent certain variables. This option also shows a plot of the squared cosines for the first three principal components.
Figure 4.12 Squared Cosines

Note: If the Sparse estimation method is used and the number of components selected is less than three, only the specified number of components are displayed in the plot.
Partial Contribution of Variables
Shows or hides a table that contains the partial contributions of variables. The partial contributions enable you to see the percentage that each variable contributes to each principal component. This option also shows a plot of the partial contributions for the first three principal components.
Figure 4.13 Partial Contribution of Variables

Note: If the Sparse estimation method is used and the number of components selected is less than three, only the specified number of components are displayed in the plot.
Summary Plots
Shows or hides the summary information produced in the default report. This summary information includes a plot of the eigenvalues, a score plot, and a loading plot. By default, the report shows the score and loading plots for the first two principal components. There are options in the report to specify which principal components to plot. See “Principal Components Report”.
Tip: Select the tips of arrows in the loading plot to select the corresponding columns in the data table. Hold down Control and click on an arrow tip to deselect the column.
Biplot
Shows or hides a plot that overlays the Score Plot and the Loading Plot for the specified number of components.
Figure 4.14 Biplot

Note: The score plot markers are dots and the loading plot markers are diamonds.
Scatterplot Matrix
Shows or hides a matrix of score and loading plots for a specified number of principal components. The scatterplot matrix arranges both the score plots and the loading plots in one space. The score plots have a yellow shaded background. The loading plots have a blue shaded background.
Figure 4.15 Scatterplot Matrix

Note: The loading plot matrix displayed in the Scatterplot Matrix is the transpose of the loading plot matrix that you obtain when you select the Loading Plot option.
Scree Plot
Shows or hides a graph of the eigenvalue for each component. This scree plot helps in visualizing the dimensionality of the data space.
Figure 4.16 Scree Plot

Score Plot
Shows or hides a matrix of scatterplots of the scores for pairs of principal components for the specified number of components. This plot is shown in Figure 4.4 (left-most plot).
Loading Plot
Shows or hides a matrix of two-dimensional representations of factor loadings for the specified number of components. The loading plot labels variables if the number of variables is 30 or fewer. If there are more than 30 variables, the labels are off by default. This information is shown in Figure 4.4 (right-most plot).
Tip: Select the tips of arrows in the loading plot to select the corresponding columns in the data table. Hold down Ctrl and click on an arrow tip to deselect the column.
Score Plot with Imputation
(Not available for the Wide or Sparse estimation methods.) Imputes any missing values and creates a score plot. This option is available only if there are missing values.
3D Score Plot
(Not available for the Wide or Sparse estimation methods.) Shows or hides a 3D scatterplot of any three principal component scores. When you first invoke the command, the first three principal components are presented.
Figure 4.17 Scatterplot 3D Score Plot

The variables show as rays in the plot. These rays, called biplot rays, approximate the variables as a function of the principal components on the axes. If there are only two or three variables, the rays represent the variables exactly. The length of the ray corresponds to the eigenvalue or variance of the principal component.
Display Options
Enables you to show or hide arrows on all plots that can display arrows. Arrows are shown if the number of variables is 1000 or fewer. If there are more than 1000 variables, the arrows are off by default.
Arrow Lines
Shows or hides arrow lines for the analysis variables in the all plots that can display arrows.
Show Supplementary Variable
(Available only if you specify a supplementary variable.) Shows or hides the arrow lines for the supplementary variables in the biplot and loading plots.
Factor Analysis
(Not available for the Wide or Sparse estimation methods.) Performs factor analysis-style rotations of the principal components, or factor analysis. See the Factor Analysis chapter in the Consumer Research book for details.
(Not available for the Wide or Sparse estimation methods.) Performs a cluster analysis on the variables by dividing the variables into non-overlapping clusters. Variable clustering provides a method for grouping similar variables into representative groups. Each cluster can then be represented by a single component or variable. The component is a linear combination of all variables in the cluster. Alternatively, the cluster can be represented by the variable identified to be the most representative member in the cluster. See the “Cluster Variables” chapter for details.
Note: Cluster Variables uses correlation matrices for all calculations, even when you select the on Covariance or on Unscaled options.
Figure 4.18 Cluster Summary

Save Principal Components
Saves the number of principal components that you specify to the data table with a formula for computing each component. The formula cannot evaluate rows with missing values.
The calculation for the principal components depends on which matrix you select for extraction of principal components:
‒ For the on Correlations option, the ith principal component is a linear combination of the centered and scaled observations using the entries of the ith eigenvector as coefficients.
‒ For the on Covariances options, the ith principal component is a linear combination of the centered observations using the entries of the ith eigenvector as coefficients.
‒ For the on Unscaled option, the ith principal component is a linear combination of the raw observations using the entries of the ith eigenvector as coefficients.
Note: If the specified number of components exceeds the rank of the correlation matrix, then the number of components saved is set to the rank of the correlation matrix.
Save Predicteds
Saves the predicted variables with a specified number of principal components to new columns in the data table.
Save DModX
Saves the observation distance to the principal components model (DModX) to a new column in the data table.DModX is defined as follows:

DModX is calculated based on the residuals,  , the number of variables, K, and the number of principal components, A. Larger DModX values indicate mild to moderate outliers in the data.
, the number of variables, K, and the number of principal components, A. Larger DModX values indicate mild to moderate outliers in the data.
Save Individual Squared Cosines
Saves the individual squared cosines to new columns in the data table.
Save Individual Partial Contributions
Saves the individual partial contributions to new columns in the data table.
Save Rotated Components
(Not available for the Wide or Sparse estimation methods.) Saves the rotated components to the data table, with a formula for computing the components. This option is available only after the Factor Analysis option is used. The formula cannot evaluate rows with missing values.
Save Principal Components with Imputation
(Not available for the Wide or Sparse estimation methods.) Imputes missing values, and saves the principal components to the data table. The column contains a formula for doing the imputation and computing the principal components. This option is available only if there are missing values.
Save Rotated Components with Imputation
(Not available for the Wide or Sparse estimation methods.) Imputes missing values and saves the rotated components to the data table. The column contains a formula for doing the imputation and computing the rotated components. This option is available only after the Factor Analysis option is used and if there are missing values.
Creates a specified number of principal component formulas and saves them as formula column scripts in the Formula Depot platform. If a Formula Depot report is not open, this option creates a Formula Depot report. See the Formula Depot chapter in the Predictive and Specialized Modeling book.
See the JMP Reports chapter in the Using JMP book for more information about the following options:
Local Data Filter
Shows or hides the local data filter that enables you to filter the data used in a specific report.
Redo
Contains options that enable you to repeat or relaunch the analysis. In platforms that support the feature, the Automatic Recalc option immediately reflects the changes that you make to the data table in the corresponding report window.
Save Script
Contains options that enable you to save a script that reproduces the report to several destinations.
Save By-Group Script
Contains options that enable you to save a script that reproduces the platform report for all levels of a By variable to several destinations. Available only when a By variable is specified in the launch window.
..................Content has been hidden....................
You can't read the all page of ebook, please click here login for view all page.
