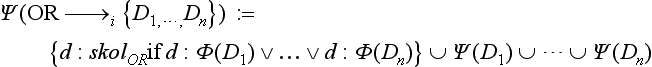A Model-Based Mediator System for Scientific Data Management
A database mediator system combines information from multiple existing source databases and creates a new virtual, mediated database that comprises the integrated entities and their relationships. When mediating scientific data, the technically challenging problem of mediator query processing is further complicated by the complexity of the source data and the relationships between them. In particular, one is often confronted with complex multiple-world scenarios in which the semantics of individual sources, as well as the knowledge to link them, require a deeper modeling than is offered by current database mediator systems. Based on experiences with federation of brain data, this chapter presents an extension called model-based mediation (MBM). In MBM, data sources export not only raw data and schema information but also conceptual models (CMs), including domain semantics, to the mediator, effectively lifting data sources to knowledge sources. This allows a mediation engineer to define integrated views based on (1) the local CMs of registered sources and (2) auxiliary domain knowledge sources called domain maps (DMs) and process maps (PMs), respectively, which act as sources of glue knowledge. For complex scientific data sources, semantically rich CMs are necessary to represent and reason with scientific rationale for linking a wide variety of heterogeneous experimental assumptions, observations, and conclusions that together constitute an experimental study. This chapter illustrates the challenges using real-world examples from a complex neuroscience integration problem and presents the methodology and some tools, in particular the knowledge-based integration of neuroscience data (KIND) mediator prototype for model-based mediation of scientific data.
12.1 BACKGROUND
Seamless data access and sharing, handling of large amounts of data, federation and integration of heterogeneous data, distributed query processing and application integration, data mining, and visualization are among the common and recurring broad themes of scientific data management. A main stream of activity in the bioinformatics domain is concerned with sequence and structural databases such as GenBank, the Protein Data Bank (PDB), and Swiss-Prot, and much work is devoted to algorithmic challenges stemming from problems (e.g., efficient sequence alignment and structure prediction). However, in addition to the well-known challenges of bioinformatics applications such as algorithmic complexity and scalability (e.g., in genomics), there are other major challenges that are sometimes overlooked, particularly when considering scientific data beyond the level of sequence and protein data (e.g., brain imagery data). These challenges arise in the context of information integration of scientific data and have to do with the inherent semantic complexity of (1) the actual source data and (2) the glue knowledge necessary to link the source data in meaningful ways. Traditional federated database system architectures, and those of the more recent database mediators developed by the database community, need to be extended to handle adequately information integration of complex scientific data from multiple sources. This extension is a combination of knowledge representation and mediator technology. In a nutshell:
Model-Based Mediation = Database Mediation + Knowledge Representation
With respect to their semantic heterogeneity (ignoring syntactic and system aspects), information integration/mediation scenarios (scientific or otherwise) can be roughly classified along a spectrum as follows: On one end, there are simple one-world scenarios; somewhere in the middle are simple multiple-world scenarios; and at the other end of the spectrum are complex multiple-world scenarios. An example of a simple one-world scenario (i.e., in which the modeled real-world entities can be related easily to one another and come from a single domain) is comparison shopping for books. A typical query is to find the cheapest price for a given book from a number of sources such as amazon.com and bn.com. An example of a simple multiple-world scenario is the integration of realtor and census data to annotate and rank real estate by neighborhood quality. Here, the approach combines and relates quite different kinds of information, but the relations between the multiple worlds are simple enough to be understood without deep domain knowledge. Examples of complex multiple-world scenarios are often found in scientific data management and are the subject of this chapter. Thus, simple and complex here refer to the degree in which specific domain semantics is required to formalize or even state meaningful associations and linkages between data objects of interest; it does not mean that the database and mediation technology for realizing such mediators is simple.1 For example, to state the problem of what the result of an integrated comparison shopping view should be, a basic understanding of a books schema (title, authors, publisher, price, etc.) is sufficient. In particular, the association operation that links objects of interest across sources can be executed (at least in principle) as a syntactic join on the ISBN. Similarly, in the realtor example, data can be joined based on the ZIP code, latitude and longitude, or street address (i.e., by spatial joins that can be modeled as atomic function calls to a spatial oracle). To understand the basic linkage of information objects, no insight into the details of the spatial join is required.
This is fundamentally different for complex multiple-world scenarios as found in many scientific domains. There, even if data is stored in state-of-the-art (often Web accessible) databases, significant domain knowledge is required to articulate meaningful queries across disciplines (or within different micro-worlds of a single discipline); further examples are offered in the next section.
Outline
In this chapter, these challenges are illustrated with examples from ongoing collaborations with users and providers of scientific data sets, in particular from the neuroscience domain (see Section 12.2). Then a methodology called model-based mediation, which extends current database mediator technology by incorporating knowledge representation (KR) techniques to create explicit representations of domain experts’ knowledge that can be used in various ways by mediation engineers and by the MBM system itself, is presented in Section 12.3. The goal of MBM could be paraphrased as:
Section 12.4 introduces some of the KR formalisms (e.g., for domain maps and process maps) and describes their use in MBM. In Section 12.5 the KIND mediator prototype and other tools being developed at the San Diego Supercomputer Center (SDSC) and the University of California at San Diego (UCSD) are presented primarily in the context of the neuroscience domain. Section 12.6 discusses related work and concludes the chapter.
12.2 SCIENTIFIC DATA INTEGRATION ACROSS MULTIPLE WORLDS: EXAMPLES AND CHALLENGES FROM THE NEUROSCIENCES
Some of the challenges of scientific data integration in complex multiple-world scenarios are illustrated using examples that involve different neuroscience worlds. Such examples occur regularly when trying to federate brain data across multiple sites, scales, and even species [3] and have led to new research and development projects aimed at overcoming the current limitations of biomedical data sharing and mediation [4].
Example 12.2.1 (Two Neuroscience Worlds).: Consider two neuro-science laboratories, SYNAPSE and NCMIR2, that perform experiments on two different brain regions. The first laboratory, SYNAPSE, studies dendritic spines of pyramidal cells in the hippocampus. The primary schema elements are thus the anatomical entities reconstructed from 3D serial sections. For each entity (e.g., spines, dendrites), researchers make a number of measurements and study how these measurements change across age and species under several experimental conditions.
In contrast, the NCMIR laboratory studies a different cell type, the Purkinje cells of the cerebellum. They inspect the branching patterns from the dendrites of filled neurons and the localization of various proteins in neuron compartments. The schema used by this group consists of a number of measurements of the dendrite branches (e.g., segment diameter) and the amount of different proteins found in each of these subdivisions. Assume each of the two schemas has a class C with a location attribute that has the value Pyramidal Cell dendrite and Purkinje Cell, respectively.
How are the schemas of SYNAPSE and NCMIR related? Evidently, they carry distinctly different information and do not even enter the purview of the schema conflicts usually studied in databases [5]. To the scientist, however, they are related for the following reason: Like pyramidal neurons, Purkinje cells also possess dendritic spines. Release of calcium in spiny dendrites occurs as a result of neurotransmission and causes changes in spine morphology (sizes and shapes obtained from SYNAPSE). Propagation of calcium signals throughout a neuron depends on the morphology of the dendrites, the distribution of calcium stored in a neuron, and the distribution of calcium binding proteins, whose subcellular distribution for Purkinje cells are measured by NCMIR.
Thus, a researcher who wanted to model the effects of neurotransmission in hippocampal spines would get structural information on hippocampal spines from SYNAPSE and information about the types of calcium binding proteins found in spines from NCMIR. Note that neither of the sources contains information that would allow a mediator system to bridge the semantic gap between them. Therefore, additional domain knowledge—independent of the observed experimental raw data of each source—is needed to connect the two sources. The domain expert, here a neuroscientist, it is easy to provide the necessary glue knowledge
Purkinje cells and Pyramidal cells have dendrites that have higher-order branches that contain spines. Dendritic spines are ion (calcium) regulating components. Spines have ion binding proteins. Neurotransmission involves ionic activity (release). Ion-binding proteins control ion activity (propagation) in a cell. Ion-regulating components of cells affect ionic activity (release).
To capture such domain knowledge and make it available to the system, the proposed approach employs two kinds of ontologies, called domain maps and process maps, respectively. The former are aimed at capturing the basic domain terminology, and the latter are used to model different process contexts. Ontologies, such as the domain map in Figure 12.1, are often formalized in logic (in this case statements in description logic [6]; see Section 12.4.1). Together with additional inference rules (e.g., capturing transitivity of has), logic axioms like these formally capture the domain knowledge and allow mediator systems to work with this knowledge (e.g., a concept or class hierarchy can be used to determine whether the system should retrieve objects of class C′ when the user is looking for instances of C).

FIGURE 12.1 A domain map for SYNAPSE and NCMIR (left) and its formalization in description logic (right). Unlabeled, gray edges ≈ “isa” ≈ “![]() ”.
”.
Domain maps not only provide a concept-oriented browsing and data exploration tool for the end user, but—even more importantly—they can be used for defining and executing integrated view definitions (IVDs) at the mediator. The previous real-world example illustrates a fundamental difference in the nature of information integration as studied in most of the database literature and as is necessary for scientific data management. In the latter, seemingly unconnected schema can be semantically close when situated in the scientific context, which, in this case, is the neuroanatomy and neurophysiological setting described previously. Therefore, this is called mediation across multiple worlds and it is facilitated using domain maps such as the one shown (see Figure 12.1).
12.2.1 From Terminology and Static Knowledge to Process Context
While domain maps are useful to put data into a terminological and thus somewhat static knowledge context, a different knowledge representation has to be devised when trying to put data into a dynamic or process context. Consider, for example, the groups of neuroscientists who study the science of mammalian memory and learning. Many of these groups study a phenomena called long-term potentiation (LTP) in nerve cells, in which repeated or sustained input to nerves in specific brain regions (such as the hippocampus) conditions them in such a manner that after some time, the neuron produces a large output even with a small amount of known input. Given this general commonality of purpose, however, individual scientists study and collect observational data for very different aspects of the phenomena.
Example 12.2.2 (Capturing Process Knowledge).: Consider a group [7] that studies the role of a specific protein N-Cadherin in the context of synapse formation during late-phase long-term potentiation (L-LTP), a subprocess of LTP. The data collected by the group consists of measurements that illustrate how the amount of N-Cadherin and the number of synapses (nerve junctions) both simultaneously increase in cells during L-LTP. Now consider that a different group [8] studies a new enzyme called CAMK-IV and its impact on a chemical reaction called phosphorylation of a protein called CREB. Their data are collected to show how modulating the amounts of CAMK-IV and other related enzymes affect the amount of CREB production, and how this, in turn, affects other products in the nucleus of the neurons. Ideally, the goal of mediating between experimental information from these two sources would be to produce an integrated view that enables an end-user scientist to get a deeper understanding of the LTP phenomena. Specifically, the end user should be able to ask queries (and get answers) that exploit the scientific interrelationship between these experiments. In this way, the integrated access provided by a mediator system can lead to new observations and questions, thus eventually driving new experiments.
At the risk of oversimplification, the first group looks at synapse formation and is only interested in the fact that some proteins (including N-Cadherin) bring about the formation of synapses. They do not look at the processes leading to the production of these proteins. The second group looks at a specific chain of events leading up to the production of the proteins but does not identify which proteins are produced. The semantic connection between these two sources can be constructed in terms of the underlying event structure and the way the two groups elaborate on different parts of it. Figure 12.2 depicts a simplified view of the relationship explained previously and shows the cyclic progression of events leading to synapse formation during LTP: Red edges situate the first source with respect to the overall process, and blue edges situate the second source. In either case, the dashed lines show the subsequence of events the sources glossed over, or abstracted. Thus, the first source does not have any information pertaining to phosphorylates (CAMK-IV, CREB), and the second source does not have any data related to forms (protein, synapse). Neither source has any data about the (black) edge synthesizes (gene, protein).

FIGURE 12.2 A simple process map. Blue and red edges (marked b and r, respectively) depict processes about which two data sources/research groups have observational data; dashed edges indicate abstractions (short cuts). No observational data is available for the edge 6–7; hence, this edge is shown in black (unmarked).
Domain maps allow data providers to put their source data into a static/terminological context, and process maps allow them to do the same for a dynamic/process context. Together, they capture valuable glue knowledge that resides at the mediator and facilitates integration of hard-to-correlate sources: in particular, concept-oriented data discovery (semantic browsing) [9], view definition, and semantic query optimization [10]. To make model-based mediation effective, it is also necessary to hook the elements of the source schema to the domain map and the process map. This process, called the contextualization mechanism, is central to the MBM framework.
12.3 MODEL-BASED MEDIATION
In mediator systems, differences in syntax and data models of sources S1, S2, … are resolved by wrappers that translate the raw data into a common data format, typically extensible markup language (XML). In most current mediator systems, all other differences, in particular schema heterogeneities, are then handled by an appropriate integrated view definition (IVD), which is defined using an XML query language [11, 12]. This architecture is extended by lifting exported source data from the level of uninterpreted, semistructured data in XML syntax to the semantically rich level of conceptual models (CMs) with domain knowledge. Then, the mediator’s views can be defined in terms of CMs (i.e., IVDs are defined in a global-as-view fashion) and thus make use of a semantically richer model involving class hierarchies, complex object structure, and properties of relationships (relational constraints, cardinalities).
12.3.1 Model-Based Mediation: The Protagonists
The underlying methodology and procedures of MBM involve users in different roles and at different levels:
![]() Data providers are typically domain experts, such as bench scientists who would like to make their data from experimental studies available to the community. In MBM, data providers can not only export an XML-queriable version of their data, but they can also export domain semantics by lifting the exported data and schema information from a structural level (e.g., XML DTDs [Document Type Definitions]) to the level of CMs.3 Allowing data providers to situate or contextualize (see Example 12.3.2) their primary data themselves has significant benefits. First, data providers know best where their data fit on the glue maps. Second, even without the IVDs defined by mediation engineers, data are automatically associated across different sources via their domain/process map contexts.
Data providers are typically domain experts, such as bench scientists who would like to make their data from experimental studies available to the community. In MBM, data providers can not only export an XML-queriable version of their data, but they can also export domain semantics by lifting the exported data and schema information from a structural level (e.g., XML DTDs [Document Type Definitions]) to the level of CMs.3 Allowing data providers to situate or contextualize (see Example 12.3.2) their primary data themselves has significant benefits. First, data providers know best where their data fit on the glue maps. Second, even without the IVDs defined by mediation engineers, data are automatically associated across different sources via their domain/process map contexts.
![]() View providers specify integrated view definitions (IVDs), that is, they program complex views in an expressive, declarative rule language. The IVDs are defined over the registered complex sources CM (S1), CM (S2), … and the glue knowledge sources in the mediator’s repository. Thus, view providers are the actual mediation engineers and they bring together (as a team or individually) expertise in the application domain and in databases and knowledge representation.
View providers specify integrated view definitions (IVDs), that is, they program complex views in an expressive, declarative rule language. The IVDs are defined over the registered complex sources CM (S1), CM (S2), … and the glue knowledge sources in the mediator’s repository. Thus, view providers are the actual mediation engineers and they bring together (as a team or individually) expertise in the application domain and in databases and knowledge representation.
The new fused objects defined by an IVD can be contextualized, based on the contexts provided by the source conceptual models (see right side of Figure 12.6). In this way, an integrated, virtual view exported by the mediator becomes a first-class citizen of the federation; it is considered a conceptual level source CM (M) itself and can be used just like any original CM-wrapped source.

FIGURE 12.6 Top: Architecture of the KIND model-based mediator. Bottom: Snapshot of the prototype. Background left shows a mediator shell for issuing ad hoc queries against CM(M); background right shows a generated subgraph having the requested result data shown in their anatomical context. Clicking on (diamond) result node retrieves the actual result data (see foreground center).
![]() End users can start with semantic browsing of CMs, by navigating the domain and process ontologies in the style of topic maps, in which a user navigates through a concept space by following certain relationships, going up and down concept hierarchies and so on. Users may also focus their view by issuing graph queries over domain or process maps, which return only the subgraphs of interest. Eventually, the user can access raw data from different sources, which is (due to contextualization) automatically organized by context [9], and access derived data resulting from user queries against the mediated views.
End users can start with semantic browsing of CMs, by navigating the domain and process ontologies in the style of topic maps, in which a user navigates through a concept space by following certain relationships, going up and down concept hierarchies and so on. Users may also focus their view by issuing graph queries over domain or process maps, which return only the subgraphs of interest. Eventually, the user can access raw data from different sources, which is (due to contextualization) automatically organized by context [9], and access derived data resulting from user queries against the mediated views.
12.3.2 Conceptual Models and Registration of Sources at the Mediator
The following components of the conceptual model CM of a source S can be distinguished:
CM(S) = OM(S) ∪ ONT(S) ∪ CON(S)
The different logical components and their dependencies are depicted in Figure 12.3:
![]() OM(S) is the object model of the source S and provides signatures for classes, associations between classes, and functions. OM(S) structures can be defined extensionally by facts (EDB), or intensionally via rules (IDB).
OM(S) is the object model of the source S and provides signatures for classes, associations between classes, and functions. OM(S) structures can be defined extensionally by facts (EDB), or intensionally via rules (IDB).
![]() ONT(S) is the local ontology of the source S. It defines concepts and their relationships from the source’s perspective.
ONT(S) is the local ontology of the source S. It defines concepts and their relationships from the source’s perspective.
![]() ONTG(S) is the ontological grounding of OM(S) in ONT(S), that is, a mapping between the object model OM(S) (classes, attributes, associations) and the concepts and relationships of ONT(S).
ONTG(S) is the ontological grounding of OM(S) in ONT(S), that is, a mapping between the object model OM(S) (classes, attributes, associations) and the concepts and relationships of ONT(S).
![]() CON(S) is the contextualization of the local source ontology relative to a mediator ontology, ONT(M).
CON(S) is the contextualization of the local source ontology relative to a mediator ontology, ONT(M).
![]() IVD(M) is the mediator’s integrated view definition and comprises logic view definitions in terms of the sources’ object models OM(S) and the mediator’s ontology ONT(M). By posing queries against the mediator’s IVD(M), the user has the illusion of interacting with a single, semantically integrated source instead of interacting with independent, unrelated sources.
IVD(M) is the mediator’s integrated view definition and comprises logic view definitions in terms of the sources’ object models OM(S) and the mediator’s ontology ONT(M). By posing queries against the mediator’s IVD(M), the user has the illusion of interacting with a single, semantically integrated source instead of interacting with independent, unrelated sources.
In the following, the local parts of CM(S) (OM(S), ONT(S), and ONTG(S)) are presented through a running example. For details on the contextualization CON(S) see Example 12.3.2 and the related work on registering scientific data sources [13].
Example 12.3.1 (Cell-Centered Database [CCDB]).: Figure 12.4 shows pieces of a simplified version of the conceptual model CM(CCDB) of a real-world scientific information source called the Cell-Centered Database [14]. The database consists of a set of EXPERIMENTS objects. Each experiment collects a number of cell IMAGES from one or more instruments. For each image, the scientists mark out cellular STRUCTURES in the image and perform measurements on them [14]. They also identify a second set of regions, called DEPOSITS, in images that show the deposition of molecules of proteins or genetic markers. In general, a region marked as deposit does not necessarily coincide with a region marked as a structure.

FIGURE 12.4 Conceptual model for registering the Cell-Centered Database [14].
Note that OM(CCDB) in Figure 12.4 includes classes that are instantiated with observed data, that is, the extensional database EDB (CCDB). In addition to classes, OM (CCDB) stores associations, which are n-ary relationships between object classes. The association co_localizes_with specifies which pairs of substances occur together in a specific structure. The object model also contains functions, such as the domain specific methods that can be invoked by a user as part of a query. For example, when the mediator or another client calls the function CCDB. deposit_in_structure ( ), and supplies the ID of a deposit object, the function returns a set of structure objects that spatially overlap with the specified deposit object.
Next, the source’s local ontology, ONT (CCDB) is described. Here, an ontology ONT (S) consists of a set of concepts and inter-concept relationships,4 possibly augmented with additional inference rules and constraints.5 The ontological grounding ONTG (S) links the object model OM (S) to the source ontology ONT (S). The source ontology serves a number of different purposes.
Creating a Terminological Frame of Reference
For defining the terminology of a specific scientific information source, the source declares its own controlled vocabulary through ONT (S). More precisely, ONT (S) comprises the terms (i.e., concepts) of this vocabulary and the relationships among them. The concepts and relationships are often represented as nodes and edges of a directed graph, respectively. Two examples of inter-concept relations are has (co) and has (pm), which are different kinds of part-whole relationships.6 In Figure 12.4, items ONT1 and ONT2 show fragments of such a concept graph. Once a concept graph is created for a source, one may use it to define additional constraints on object classes and associations.
Semantics of Relationships
The edges in the concept graph of the source ontology represent inter-concept relationships. Often these relationships have their own semantics, which must be specified within ONT (S). Item ONT4 declares two new relationships, tc_has (co) and tc_has (pm). After registration, the mediator interprets this declaration and creates the new (possibly materialized) transitive relations on top of the base relations has (co) and has (pm) provided by the source S. Similarly, the item ONT5 is interpreted by the mediator using a higher-order rule for chaining binary relations:
![]()
With this, ONT5 creates a new relationship has_co_pm (X, Y) provided that there is a Z such that tc_has (co) (x, Z), and tc_has (pm) (Z, Y).
Ontological Grounding of OM (S)
A local domain constraint specifies additional properties of the given extensional database and thereby establishes an ontological grounding ONTG (S) between the local ontology ONT (S) and the object model OM (S) (see Figure 12.3). Items OG1-OG2 in Figure 12.4 refine the domains of the attributes EXPERIMENT. cell_type and STRUCTURE. name from the original type declaration (STRING). The refinement constrains them to take values from those nodes of the concept graph that are descendants of the concept cerebellum through the has (co) relationship.
This constraint illustrates an important role of the local ontology in a conceptually lifted source. By constraining the domain of an attribute to be concept name, C, the corresponding object instance o is semantically about C. In addition, this also implies that o is about any ancestor concept, C′, of C where ancestor is defined via has (co) edges only. Similarly, if a specific instance, STRUCTURE. name, has the value spine process, it is also about dendrite (ONT2 in Figure 12.4).
In addition to linking attributes to concept names, a constraint may also involve inter-concept relationships. Assume projects_to (cell, brain_region) is a relationship in the source ontology ONT(CCDB). A constraint may assert that for all instances e of class EXPERIMENT, projects_to (e.cell_type, ‘globus_pallidus’) holds (0G3). The constraint thus refines the original relationship projects_to to suit the specific semantics of OM(CCDB). Such constraint-defined correspondences between OM (S) and ONT (S) are used in the contextualization process [13].
Intensional Definitions
In the CM wrapper of a source, S, one can define virtual classes and associations that can be exported to the mediator as first-class, queriable items by means of an intensional database IDB (S). For example, one can create a new virtual class called DENATURED_PROTEIN in IDB (CCDB) via the rule:
![]()
Thus, an instance of a DENATURED_PROTEIN is created when a dark protein deposit is recorded in an instance of DEPOSIT and there is some structure in which this deposit is found. As a general principle of creating a CM wrapper, such a definition will be supplemented by additional constraints to connect it to the local ontology. For example, assume that ONT (CCDB) already contains a concept called process. Item ONT3 defines denaturation as a specialization of process. The constraint OG4 completes the semantic specification about the new DENATURED_PROTEIN object.
Contextual References
It is a common practice for scientific data sources to tag object instances with attributes from a public standard and to use controlled vocabularies for the values of some of these attributes. For example, the source can specify that the domain of the DEPOSIT. id field can be accessed through an internal method, which, given a protein name, gets its id from a specific database. For example, one can use get_expasy_protein_id to retrieve this information from the Swiss-Prot database on the Web. How the source enforces this integrity constraint is internal to the source and not part of its conceptual export schema.
12.3.3 Interplay Between Mediator and Sources
To address the source registration issue, which components of an existing n-source federation that can be seen, or accessed, by the new, n+1st source need to be specified. A federation at the mediator consists of: (1) currently registered conceptual models CM (S) of each participating source S, (2) one or more global ontologies ONT (M)residing at the mediator that have been used in the federation, and (3) integrated views IVD (M) defined in a global-as-view (GAV) fashion.
Typical mediator ontologies ONT (M) are public, meaning they serve as domain-specific expert knowledge and thus can be used to glue conceptual models from multiple sources. Examples of such ontologies are the Unified Medical Language System (UMLS) from the National Library of Medicine7 and the Biological Process Ontology from the Gene Ontology Consortium.8 In the presence of multiple ontologies, articulations, (mappings between different source ontologies [16]) can be used to register with the mediator information about inter-source relationships. Note that a source, S, usually cannot see all of the previously discussed components (1–3) when defining its conceptual model: Although S sees the mediator’s ontologies, ONT (M), and thus can define its own conceptual model, CM (S), relative to the mediator’s ontology in a local-as-view (LAV) fashion, it cannot directly employ another source’s conceptual model, CM (S‘), nor can it query the mediator’s integrated view, IVD (M), which is defined global-as-view (GAV) on top of the sources. The former is no restriction because S’ can register CM (S′), in particular ONT (S’), with the mediator, at which point S can indirectly refer to registered concepts of S‘ via ONT (M). The latter guarantees that query processing in this setting does not involve recursion through the Web (i.e., between a source S and the mediator M). The dependency graph in Figure 12.3 is acyclic.9
Example 12.3.2 (Contextualization: Local-as-View).: Consider the domain map in Figure 12.5. Lighter-colored nodes correspond to concepts that the mediator understands and a source can see. Now assume a source, S, wants to register information about specific neurons and their dendrites, but the mediator ontology (domain map) does not have dedicated names for those specific kinds of neurons and dendrites. In MBM this problem is solved by contextualizing the new local source concepts as views on the mediator’s global concepts: In Figure 12.5, the darker-colored source concepts are hooked to the mediator’s domain map, thereby defining their meaning relative to the mediator’s concepts. This is achieved by sending the following first-order axioms (here in description logic syntax) to the mediator:

Thus instances of MyDendrite are exactly those dendrites that express Dopamine R(eceptor), and MyNeuron objects are medium spiny neurons projecting to Globus Pallidus External and only have MyDendrites. Assuming properties are inherited along the transitive closure of isa, it follows that MyNeuron, like any Medium_Spiny_Neuron projects to certain structures (OR in Figure 12.5). With the newly registered knowledge, it follows that MyNeuron definitely projects to Globus-Pallidus-External. To specify that it only projects to the latter, a nonmonotonic inheritance (e.g., using F-logic with well-founded semantics) can be employed.
Note that the intuitive graphical contextualization depicted in Figure 12.5 is not unique; logically equivalent domain maps may have different graphical representations.10 For domain maps that can be completely axiomatized using a description logic, a reasoning system such as Fast Classification of Terminologies (FaCT) [17] can be employed to compute the deductive closure and, in particular, to derive a unique concept hierarchy and check consistency of a domain map.
12.4 KNOWLEDGE REPRESENTATION FOR MODEL-BASED MEDIATION
This section takes a closer look at the principal mechanisms for specifying glue knowledge: ontologies in the form of domain maps (DMs) and process maps (PMs).
12.4.1 Domain Maps
As is standard for ontologies, DMs name and specify relevant concepts by describing the characteristic relationships among them [18]. In this way, DMs provide the basic domain semantics needed to glue data across different sources in multiple-world scenarios. DMs can be depicted more intuitively in the form of labeled, directed graphs. In contrast to many other graph-based notations, however, DMs have a solid formal semantics via a translation to logic rules. The graph form of DMs is defined as follows.
Definition 12.1 Domain Maps
Let C be a set of symbols called concepts and ![]() a set of roles. A DM is a directed, labeled graph with nodes C. A concept C ∈ C can be understood as denoting a class of objects sharing a set of common properties. To understand how a concept C is defined relative to other concepts, one needs to inspect its outgoing edges. c ∈ C denotes that c is an instance of concept C.11 Edges are distinguished in DMs as follows:
a set of roles. A DM is a directed, labeled graph with nodes C. A concept C ∈ C can be understood as denoting a class of objects sharing a set of common properties. To understand how a concept C is defined relative to other concepts, one needs to inspect its outgoing edges. c ∈ C denotes that c is an instance of concept C.11 Edges are distinguished in DMs as follows:
1. C ![]() D (short: C → D) defines that every C isa D, that is, c ∈ C implies c ∈ D.
D (short: C → D) defines that every C isa D, that is, c ∈ C implies c ∈ D.
The subconcept/subclass relation is very common in DMs, thus the isa label is usually omitted and the shorthand notation C → D is used instead.
2. C ![]() D defines that for every c ∈ C, there exists some r-related d ∈ D. Here, r ∈
D defines that for every c ∈ C, there exists some r-related d ∈ D. Here, r ∈ ![]() is a role, or, a binary relation r(c, d) between instances of C and D
is a role, or, a binary relation r(c, d) between instances of C and D
3. C ![]() D defines that for every c ∈ C and all x that are r-related to c (i.e., for which r(c, x) holds), x ∈ D holds.
D defines that for every c ∈ C and all x that are r-related to c (i.e., for which r(c, x) holds), x ∈ D holds.
4. C ![]() D defines that if c ∈ C and d ∈ D, then they are r-related, that is, r(c, d) holds.
D defines that if c ∈ C and d ∈ D, then they are r-related, that is, r(c, d) holds.
5. AND→i{D1, …, Dn} indicates that an AND-node with n outgoing edges to D1, …, Dn, respectively, defines an anonymous concept, the intersection of concepts D1, …, Dn.
6. OR→i{D1, …, Dn}, indicates that an OR-node with n outgoing edges to D1, …, Dn, respectively, defines an anonymous concept, the union of concepts D1, …, Dn.
7. C ![]() D defines that C is equivalent to D, meaning every C isa D and vice versa. It could have been denoted also as C ↔ D. However, the directed edge keeps the distinction between C (the definiendum) and its definition D (definiens).
D defines that C is equivalent to D, meaning every C isa D and vice versa. It could have been denoted also as C ↔ D. However, the directed edge keeps the distinction between C (the definiendum) and its definition D (definiens).
Note that D can be an atomic or a defined concept. When unique, AND nodes are omitted and outgoing arcs directly attached to the concept being defined. In Figure 12.5, unlabeled, grey edges and edges labeled proj (projects-to) correspond to isa edges and ex:proj edges, respectively.
Reified Roles as Concepts
In DMs, as in description logics, the concepts are being defined, whereas the roles are only a means to that end. To capture the semantics of roles, or define their properties in terms of each other, they need to be defined in terms of concepts themselves. In logic, this “quoting mechanism” is known as reification.
Example 12.4.1 (Roles as Concepts).: Consider a DM involving the roles regulates, activates, and inhibits, and assume that in the given domain, activates (C, D) and inhibits (C, D) are special cases of regulates (C, D). Instead of introducing a special notation for sub-roles12 and then defining the mechanics of how roles can be related to one another, roles are turned into first-class citizens by making them concepts using an operator, make-concept (mc). The modeling capabilities of DMs can be applied to roles and, for example, simply state that mc(activates) ![]() mc(regulates).
mc(regulates).
By modeling roles as concepts, more domain semantics can be formalized, leading to better knowledge engineering. In particular, during query processing, such formalized knowledge can be automatically employed by the system: Given a DM (formalized as logic rules), an MBM query or view definition involving activates and regulates knows that the former is a subconcept of the latter. If during query processing a goal regulates (‘cAMP’, Protein) is evaluated, the logic rules corresponding to the DM knowledge allow the system to deduce that any result for activates (‘cAMP’, Protein) is also an answer for regulates (‘cAMP’, Protein). This is correct because a substitutability principle holds, which allows the system to replace a concept, D, with any of its subconcepts, C, that is, for which C ![]() D holds.
D holds.
Generating the Role Hierarchy
When making a role into a concept, the isa hierarchy13 on concepts induces an isa hierarchy on roles.
Domain Maps as Logic Rules
Domain maps borrow from description logics [19] the notions of concept and roles. Indeed, while some of the previously mentioned constructs of DMs have equivalent formalizations in description logic [20], the fact that additional mechanisms are needed such as roles as concepts and recursive and parameterized roles and concepts, and the fact that executable DMs are wanted during query processing, require a translation into a more general logic framework.
In the following, DMs are formalized in a minimal subset of F-logic [21]. The semantics of DMs could be formalized in other languages, in particular in other deductive database languages. The use of F-logic is convenient because a small subset of it already matches nicely the minimal requirements established for a MBM system [20]. Moreover, implementations of F-logic are readily available [22, 23] and have been used by the authors in different mediator prototypes before [24, 25, 26].
In F-logic, c : C and C :: D denote class membership (c ∈ C) and subclassing (C ⊆ D), respectively. Thus, there are logic rules of the form head if body that express the F-logic semantics of “:” and “::” Say that “::” is a reflexive, transitive, and antisymmetric14 relation.
Definition 12.2 Compilation of Domain Maps
The mapping Ψ : DM → FL of domain maps to F-logic is defined as follows:
Remarks
Here, Φ(D) is defined similar to Ψ(D), but it returns for a compound concept description D, a new auxiliary symbol Φ(D) representing the compound. For atomic D, Φ(D) = Ψ(D) holds simply. The symbols skolx produce new Skolem function symbols every time they are used in the translation Ψ: For example, in 4(a), a symbolic representation is invented for the existentially quantified variable_d. Note that c, d, _d are logic variables, while C, D, Di, and False are constants.15 The different variants (a) and (b) in the translations of DMs correspond to different intended uses: in 4(a), an anonymous object is created for the ∃-quantified variable, in 5(a), all C.r objects are type coerced into instances of D. In contrast, the (b) translations only check whether the constraints induced by the DM edges are indeed satisfied and signal an inconsistency (False) if they are not.
Example 12.4.2 Roles as Concepts Continued.: Consider a DM stating that NProt ![]() Protein, NProt regulates some Gene, and cfos
Protein, NProt regulates some Gene, and cfos ![]() Gene.16 The role regulates is conceptualized by asserting mc(regulates). When making its hidden arguments visible, mc (regulates (C, D)) really denotes a family of regulates concepts. The isa hierarchy on regulates concepts is derived from the isa hierarchy of its arguments. For example:
Gene.16 The role regulates is conceptualized by asserting mc(regulates). When making its hidden arguments visible, mc (regulates (C, D)) really denotes a family of regulates concepts. The isa hierarchy on regulates concepts is derived from the isa hierarchy of its arguments. For example:
![]()
Deriving the Role Hierarchy
Previously the unary operator, mc, which turns role literals into concepts was introduced. It is implemented in FL as a subclass of the (meta-)class concept by asserting mc : : concept and adding further rules for deriving the role hierarchy from the concept hierarchy, which are given as set of mc-declarations such as r(C, D): mc by the user:

Observe that with these rules, the desired result is obtained in Example 12.4.2.
Recursive Concepts
Consider the part of relationship has_a and its interaction with isa. For example, MyNeuron isa Medium_Spiny_Neuron, which in turn has_a Neostriatum therefore MyNeuron has_a Neostriatum holds (see Figure 12.5). In the general case, this gives rise to a recursive rule if C ![]() D and D
D and D ![]() E then C
E then C ![]() E. Similarly, one can define that isa and has_a are independently transitive and that isa is anti-symmetric. For such recursive definitions, an intuitive graph notation can be devised (e.g., using a dashed edge for the concept being defined to its recursive definition, see Ludäscher et al. [27]). In a declarative, rule-based query language like F-logic, an executable specification is:
E. Similarly, one can define that isa and has_a are independently transitive and that isa is anti-symmetric. For such recursive definitions, an intuitive graph notation can be devised (e.g., using a dashed edge for the concept being defined to its recursive definition, see Ludäscher et al. [27]). In a declarative, rule-based query language like F-logic, an executable specification is:
![]()
Note that x, Y, Z are concept variables. Such F-logic rules can also be used at the mediator to handle inductive definitions, such as ONT4 in Figure 12.4, in particular, when the source does not have the capability to evaluate recursive definitions.
Parameterized Roles and Concepts
Part of relationships such as has_a come in different flavors, F (e.g., F ∈ {member/collection, portion/mass, phase/activity, …}) and transitivity does not necessarily carry over across flavors [15].17 This is most naturally modeled by a parameterized role, has_a (F), which is transitive within each flavor, F, but which may interact in other ways across flavors. Definition 12.2 shows how domain maps can be formalized as logic rules via a mapping Ψ. This mapping can be extended for parameterized roles and concepts: For example, assume the parameterized role has_a (F) should hold between concepts C and D only for some flavors, F, satisfying a condition ϕ(F). One can extend Ψ and compile such a parameterized DM edge into F-logic as follows:
Note that a parameterized role such as has_a (F) has a first-order semantics in F-logic despite its higher-order syntax [28].
12.4.2 Process Maps
PMs provide abstractions of process knowledge, that is, temporal and/or causal relationships between events that can be used for situating and linking data across different sources. Like DMs, PMs are directed, labeled graphs, albeit with a very different semantics: Nodes are used to model states and edges correspond to state transitions, which are labeled with a process name describing the transition. In this way, data providers (e.g., bench scientists) can not only hook their raw data to the (given or refined) DMs but also to processes witnessed in their experimental studies databases (see Figure 12.2 and Figure 12.8).
Initial Process Semantics PM0
Intuitively, an edge of the form ![]() of a PM means that the process Π leads from state s to s′; φ is a necessary precondition that must hold in s for π to happen, and ψ is a postcondition, which holds in s’ as a result of Π. P M0 denotes the set of all initial process semantics.
of a PM means that the process Π leads from state s to s′; φ is a necessary precondition that must hold in s for π to happen, and ψ is a postcondition, which holds in s’ as a result of Π. P M0 denotes the set of all initial process semantics.
The edge eπ of a PM is called a process occurrence of π in PM. Thus, a process occurrence specifies where in a PM a process occurs, and which pre- and postconditions, φ and ψ, this occurrence satisfies. In addition to the semantics implied by the occurrence of eΠ in PM, a process π can have an initial semantics associated with the process name, π.
To allow for parameterization of processes, edge labels where process names are first-order atoms (of the form π = π (T1, …, Tn) where each term Ti is a logic variable or constant) are considered. For example, consider π = opens (Channel) as describing the opening process of an ion channel. Its initial semantics are defined by the expression:
![]()
meaning that any transition along a process occurrence of π = opens (Channel) in a PM must be from a state where open (Channel) was false. In the successor state, however, (after π has happened), open (Channel) is true.
From Process Maps to Domain Maps
The first-order predicates occurring in φ and ψ are called open(Channel), fluents, because their truth is state dependent. It is required that the set of fluent predicate symbols, F, is disjointed from the set, P, of process names and the sets of concept and role names C and rscr;, respectively. In contrast, the constant parameters used in process occurrences, such as Channel are allowed to be concepts from C.
For example, a DM may have that NMDA_receptor ![]() Calcium_channel
Calcium_channel ![]() channel in which case the process knowledge about the opening of channels and the static knowledge from a DM are directly linked through the common concept Channel.
channel in which case the process knowledge about the opening of channels and the static knowledge from a DM are directly linked through the common concept Channel.
Similarly, just as roles are first-class citizens by reifying them into concepts, the same can be done for processes, by specifying additional semantics of processes using domain maps.
Example 12.4.3 (Processes as Concepts).: Consider the binds_to (X, Y) process with the initial semantics.
![]()
Now consider a DM in which processes were reified as concepts as follows:
![]()
It is easy to see that this (parameterized) DM edge, when translated into F-logic, allows the system to conclude in the combined knowledge base (DM ∪ PM0) that
![]()
Process Elaboration and Abstraction
The edge, eπ, of a process occurrence can be seen as an abstraction of a real process. In addition to its initial semantics, PM0, and the semantics induced by its concrete occurrence in a specific PM, this abstraction can be elaborated by replacing the eπ with a (sub-)process map elab (eπ), whose initial and final states are s and s‘. The newly created nodes and edges of the elaboration, elab (eπ), are annotated with the same unique elaboration identifier eID. The eID includes at least a reference to eΠ, indicating the edge being elaborated, and the author (data provider) of the elaboration.
The converse of elaboration, abstraction, takes a connected subgraph, π (S, s0, Sf, E), with nodes S, edges E, and distinguished nodes s0, sf ∈ S (initial and final state), and abstracts Π into a single edge eπ = abstract (Π (S, s0, sf, sf, E)). The abstracted edges E of Π are marked with a unique abstraction identifier aID, which includes a reference to the new abstraction edge, eπ, and the author of the abstraction.
Definition 12.3 Process Maps
A PM Π(S, s0, sf, E) is a connected, directed graph with nodes, S, labeled edges, E, and initial and final states s0, Sf ∈ S. The edges eπ of E are of the form
where the process name π is a first-order atom and φ and ψ are first-order formulas, called the precondition and postcondition of eΠ, respectively.
Given an edge ![]() of a process map Π(S, s0, sf, E), the elaboration, elab(e), of e is a process map Π′{S′, sa, Sb, E′) such that (1) the initial and final states are sa, Sb, (2) S′ ∩ S = {sa, Sb}, and (3) all e′ ∈ E′ are linked to e via a common, unique identifier eid(e′, e).
of a process map Π(S, s0, sf, E), the elaboration, elab(e), of e is a process map Π′{S′, sa, Sb, E′) such that (1) the initial and final states are sa, Sb, (2) S′ ∩ S = {sa, Sb}, and (3) all e′ ∈ E′ are linked to e via a common, unique identifier eid(e′, e).
A connected subgraph of a PM with distinguished initial and final state is called a subprocess map (sub-PM). Given a PM Π(S, s0, Sf, E), the abstraction of a sub-PM Π′(S‘, sa, sb, E‘) of Π, denoted abstract (Π′), is a new edge ![]() , where (i) eπ‘ ∉ E, and (ii) all e‘ ∈ E‘ are linked to eπ‘ via a common, unique identifier aid(e‘, eπ‘).
, where (i) eπ‘ ∉ E, and (ii) all e‘ ∈ E‘ are linked to eπ‘ via a common, unique identifier aid(e‘, eπ‘).
Marking edges with elaboration and abstraction identifiers guarantees one-to-one mappings between an edge and its elaboration and similarly, between a sub-PM and its abstraction. In this way, data providers can “double-click” on an edge, eπ, and elaborate the processes into a PM, Π, to provide more precise links to their data. Conversely, they may collapse a sub-PM, Π, into a single edge, eπ, if the data does not provide information at the detailed level of π and hence is more adequately hooked to the overall process, eπ.
Process Maps as Logic Rules
Similarly to DMs, one can translate PMs into a logic representation Ψ(PM). The difference is that for DMs, the formalization in description logic or F-logic yields a first-order logic semantics, whose unique minimal model, M(DM), interprets concepts and roles as unary and binary predicates over a set of individuals. The model, M, implies that data objects, which are linked as concept instances to a DM, have the properties defined by the domain map (e.g., the neurons in the images linked to MyNeuron in Example 12.3.2 project to Globus_Pallidus_External). In contrast, the logic representation of a PM specifies only some process properties via pre- and postconditions in the PM and the PM’s graph structure. The details of the semantics are omitted due to lack of space. The basic idea is that the graph structure of PMs (with its embedded hierarchy of elaborations and abstractions) is formalized via a nested Kripke structure in which the nodes of PM (states) have associated first-order models and in which labeled process edges specify a temporal accessibility relation between states.18 In particular, a process elaboration of an edge, eπ, adds to the initial semantics, PM0, and the semantics of the pre- and postconditions of the concrete occurrence of eπ in PM, an elaboration semantics (i.e., a sequence of intermediate states with first-order constraints along the paths of the elaboration).
12.5 MODEL-BASED MEDIATOR SYSTEM AND TOOLS
At the core of the MBM framework is the KIND mediator system. Other important components are the Spatial Markup and Rendering Tool (SMART) Atlas for annotating, displaying, and relating data with brain atlases, the CCDB, defined in Example 12.3.1 as the primary source of experimental data, and the Knowledge Map Explorer (Know-ME) tool for concept-based navigation of source and mediated views. For a description of Know-ME, see Qian et al. [29]; the other components are described in the following text.
12.5.1 The KIND Mediator Prototype
The architecture of the KIND mediator system is depicted on top in Figure 12.6. At the bottom, a snapshot of the prototype execution is shown: After the user issues a query against the integrated view, the system situates the results on a domain map, in this case ANATOM (simple ontology of brain anatomy). By clicking on the orange diamonds, the user can retrieve the actual result objects, grouped by concept (foreground).
In the first prototype [9, 30] the F-logic implementation FLORA [31] was used as the only query processing and deduction engine. As part of a large, collaborative project [4] the prototype is being re-implemented as a modular, distributed mediator system that includes several additional components, including the following:
![]() Logic plan generator: Given a user query, Q, and an integrated view definition IVD, Q o IVD is translated into a plan generator program PG(Q
Logic plan generator: Given a user query, Q, and an integrated view definition IVD, Q o IVD is translated into a plan generator program PG(Q ![]() IVD) that, when executed, produces an initial logic query plan for Q
IVD) that, when executed, produces an initial logic query plan for Q ![]() IVD. Here “
IVD. Here “![]() ” denotes query composition.
” denotes query composition.
![]() Query rewriter: This module takes a logic query plan and rewrites it into an executable, distributed plan based on the capabilities of a source (e.g., conjunctive queries with binding patterns or complete SQL).
Query rewriter: This module takes a logic query plan and rewrites it into an executable, distributed plan based on the capabilities of a source (e.g., conjunctive queries with binding patterns or complete SQL).
![]() Execution plan compiler: For final execution, the rewritten plan is compiled into a logic program whose run-time execution sends the corresponding sub-queries to wrapped sources, retrieves results, and post-processes them (e.g., joins, group-bys, and unions across sources) before sending them to the user.
Execution plan compiler: For final execution, the rewritten plan is compiled into a logic program whose run-time execution sends the corresponding sub-queries to wrapped sources, retrieves results, and post-processes them (e.g., joins, group-bys, and unions across sources) before sending them to the user.
![]() SQL plan generator: For relational sources (those having SQL query capabilities), this wrapper module translates a logic query plan into an equivalent SQL statement, similar to Draxler’s tool [32].
SQL plan generator: For relational sources (those having SQL query capabilities), this wrapper module translates a logic query plan into an equivalent SQL statement, similar to Draxler’s tool [32].
A preliminary version of this new system has been recently demonstrated [13] and includes all of the modules previously listed. Plan generation and rewriting is implemented using logic programming technology [33]. The SQL plan generator has been implemented in Java. It is planned that the final system will include specialized inference engines such as FLORA and XSB [34] for handling deductive and object-oriented database capabilities, and FaCT [17] for reasoning tasks over domain maps that are formalized in description logics.
12.5.2 The Cell-Centered Database and SMART Atlas: Retrieval and Navigation Through Multi-Scale Data
The CCDB mentioned earlier, in Example 12.3.1, houses different types of high-resolution, 3D light and electron microscopic reconstructions of cells and subcellular structures produced at the National Center for Microscopy and Imaging Research19 [14]. It contains structural and protein distribution information derived from confocal, multiphoton, and electron microscopy, including correlated microscopy. Many of the data sets are derived from electron tomography, a powerful technique for deriving 3D information from electron microscopic specimens. Electron tomography is similar in concept to medical imaging techniques like computerized axial tomography (CAT) scans and magnetic resonance imaging (MRI) in that it derives a 3D volume from a series of 2D projections through a structure. In this case, the structures are contained in sections prepared for electron microscopy, which are tilted through a limited angular range. Examples of datasets in the CCDB are shown on the left of Figure 12.7.

FIGURE 12.7 Left: Examples of tomographic data sets in the CCDB. A and B show a selectively stained spiny dendrite from a Purkinje cell. A is a projection of the volume reconstruction (dendrite appears as white against dark background). B is the segmented dendrite. C and D show a tomographic reconstruction of the node of Ranvier. C is a single computed slice through the volume. D is a surface reconstruction of the various components comprising the node. Scale bar in B = 1μm; in C = 0.5μm. Right: Registration of a data set with the Smart Atlas. The user draws a polygon representing the location of a data set, in this case a filled Purkinje neuron. The user specifies the database containing this data, then enters an annotation and selects a concept from the UMLS or some other ontology. The concept ID is stored in the database.
A screenshot of the Smart Atlas tool is shown on the right of Figure 12.7. It is based on a geographic mapping tool [35] and allows users to define polygons on a series of 2D vector images and annotate them with names, relationships, and concept IDs from an ontology such as UMLS. This tool provides another kind of glue map (in addition to domain and process maps). First, a brain atlas such as that by Paxinos and Watson [36] is translated into a spatial format, such as Scalable Vector Graphics (SVG). The user then marks up the atlas using the Smart Atlas tool (e.g., with concept names from UMLS). Once the atlas has been (partially) marked up, it can be queried from the same browser: Clicking on any point in the atlas will return the stereotaxic coordinates; clicking on a brain region will return the name of that region, along with any synonyms, and highlight all planes containing that structure. The Smart Atlas can now be used to register a researcher’s data to a specific spatial location. This also links the registered data automatically to the UMLS ontology by virtue of the earlier semantic markup of spatial objects. To register source data, the user draws an arbitrary polygon representing the approximate data location on one of the atlas planes (Figure 12.7, right). The user is then presented with a form that can be used to add annotations or provide additional links to concepts of an ontology. Although the UMLS is used in the examples shown here, the user will eventually be able to use multiple ontologies, including those of their own creation, for semantically indexing data. Tools are also being developed to define new terms and relationships in existing ontologies. Another component of the system has been demonstrated and shows how spatial and conceptual information can be used together in a mediator system [37]; see also Martone et al.’s chapter in Neuroscience Databases [38] for further details on the use of the Smart Atlas.
12.6 RELATED WORK AND CONCLUSION
Significant progress has been made in the general area of data mediation in recent years, and several prototype mediator architectures have been designed by projects like TSIMMIS [39], SIMS [40], Information Manifold [41], Garlic [42], and MIX [43]. While these approaches focus mostly on structural and schema aspects, the problem of semantic mediation has also been addressed: In the DIKE system [44], the focus is on automatic extraction of mappings between semantically analogous elements from different schemas. A global schema is defined in terms of a conceptual model (SDR network), in which the nodes represent concepts and the (directed) edge labels represent their semantic distances, and a score called semantic relevance measures the number of instances of the target node that are also instances of the source node. The correspondence between objects is defined in terms of synonymies, homonymies, and sub-source similarities, defined by finding maximal matching between the two graphs.
ODB-Tools [45] is a system developed on top of the MOMIS [46] system for modeling and reasoning about the common knowledge between two to-be-integrated schemas. They present the object-oriented language, ODLI3, derived from a description logic (OCDL). The language allows a user to create complex objects with finite nesting of values, union and intersection types, integrity constraints, and quantified paths. These constructs are used to define a class in one schema as a generalization, aggregation, or equivalent with respect to another; subsumption of a class by another can be inferred. An integrated schema is obtained by clustering schema elements that are close to one another in terms of an affinity metric.
Calvanese et al. [47] perform semantic information integration using a local-as-view approach by expressing the conceptual schema by a description logic language called ![]() and subsequently defining non-recursive Datalog views to express source data elements in terms of the conceptual model. The language
and subsequently defining non-recursive Datalog views to express source data elements in terms of the conceptual model. The language ![]() represents concepts, C, relations, R, and a set of assertions of the form C1 ⊂ C2 or R1 ⊂ R2, where R1, R2 are
represents concepts, C, relations, R, and a set of assertions of the form C1 ⊂ C2 or R1 ⊂ R2, where R1, R2 are ![]() relations with the same arity. Mediation is accomplished by defining reconciliation correspondences, or specifications that a query rewriter uses to match a conceptual-level term to data in different sources.
relations with the same arity. Mediation is accomplished by defining reconciliation correspondences, or specifications that a query rewriter uses to match a conceptual-level term to data in different sources.
Recently Peim et al. [48] have proposed an extension to the well-known TAMBIS system [49]. Their approach is similar to ours [18, 50] in that a logic-based ontology (in their case the ![]() description logic) interfaces with an object-wrapped source. While F-logic [28] is used here as the internal knowledge representation and query language, their work focuses on how a query on the ontology is transformed to monoid comprehensions for semantic query optimization.
description logic) interfaces with an object-wrapped source. While F-logic [28] is used here as the internal knowledge representation and query language, their work focuses on how a query on the ontology is transformed to monoid comprehensions for semantic query optimization.
12.6.2 Summary: Model-Based Mediation and Reason-Able Meta-Data
MBM was presented as a methodology that supports information integration of scientific data across complex, multiple-world scenarios as found in the neuroscience domain. In this framework, object-oriented models and conceptual models (CM), domain maps (DM), and process maps (PM) all provide means to capture more domain semantics and thus can act as glue knowledge sources to link hard-to-correlate sources. Mechanisms to contextualize source data formally were presented. The graph structures thus constructed have been shown to be useful for navigating across related concepts and querying local data during navigation [29].
Logic formalizations of DMs and PMs can be seen as “reason-able” or “executable” “meta-data” (see a paper by Horrocks [51]): Unlike conventional, descriptive meta-data, which are primarily used for data discovery, formal ontologies, such as DMs and PMs, can support much more versatile computational tasks in a mediator system, as illustrated in this chapter. For example, different and apparently unrelated data objects can be associated and retrieved together or even fused by the mediator’s integrated view definition (IVD), because IVDs can be defined as deductive rules over DMs and PMs (Figure 12.3). In this way, in model-based mediation (MBM), logic rules play the role of executable or computational meta-data for scientific data integration. The latter is a challenging application and benchmark for combined database and knowledge representation techniques.
ACKNOWLEDGMENTS
This work has been supported by NIH/NCRR 3 P41 RR08605-08S1 (Biomedical Informatics Research Network [BIRN]) and NSF-NPACI Neurosciences Thrust ACI 9619020. The authors thank their colleagues and students involved in the BIRN project for their contributions, in particular, Xufei Qian, Edward Ross, Joshua Tran, and Ilya Zaslavsky.
REFERENCES
[1] Papakonstantinou, Y., Gupta, A., Haas, L.M. Capabilities-Based Query Rewriting in Mediator Systems. Distributed and Parallel Databases. 1998;6(no. 1):73–110.
[2] Li, C., Yemeni, R., Vassalos, V., et al, Capability Based Mediation in TSIMMIS, Proceedings of the ACM International Conference on Management of Data (SIGMOD). 1998:564–566.
[3] National Partnership for Computational Infrastructure (NPACI): Neuroscience Thrust Area, 2001. http://www.npaci.edu/Thrusts/Neuro/.
[4] Biomedical Informatics Research Network Coordinating Center (BIRN-CC). University of California, San Diego, http://nbirn.net/’, 2001.
[5] Kashyap, V., Sheth, A. Semantic and Schematic Similarities Between Database Objects: A Context-Based Approach. VLDB Journal. 1996;5(no. 4):276–304.
[6] Calvanese, D., Giacomo, G.D., Lenzerini, M., et al. Description Logic Framework for Information Integration. In: Proceedings of the Sixth International Conference on Principles of Knowledge Representation and Reasoning (KR’98). Morgan Kaufmann; 1998:2–13.
[7] Bozdagi, O., Shan, W., Tanaka, H., et al. Increasing Numbers of Synaptic Puncta During Late-Phase LTP: N-Cadherin is Synthesized Recruited to Synaptic Sites and Required for Potentiation. Neuron. 2000;28(no. 1):245–259.
[8] Kasahara, J., Fukunaga, K., Miyamoto, E. Activation of Calcium/Calmodulin-Dependent Protein Kinase IV in Long Term Potentiation in the Rat Hippocampal CA1 Region. Journal of Biological Chemistry. 2001;276(no. 26):24044–24050.
[9] Heidelberg, Germany Gupta, A., Ludäscher, B., Martone, M.E., An Extensible Model-Based Mediator System with Domain Maps, Demonstration Session of the 21st International Conference on Data Engineering (ICDE). 2001.
[10] Chakravarthy, S., Grant, J., Minker, J. Logic-Based Approach to Semantic Query Optimization. ACM Transactions on Database Systems (TODS). 1990;15(no. 2):162–207.
[11] Lecture Notes in Computer Science 1777 Ludäscher, B., Papakonstantinou, Y., Velikhov, P., Navigation-Driven Evaluation of Virtual Mediated Views. Springer. Proceedings of the International Conference on Extending Database Technology (EDBT). 2000:150–165.
[12] Papakonstantinou, Y., Vassalos, V., The Enosys Markets Data Integration Platform: Lessons from the Trenches, International Conference on Information and Knowledge Management (CIKM). 2001.
[13] Lecture Notes in Computer Science 2503. Springer Gupta, A., Ludäscher, B., Martone, M.E., Registering Scientific Information Sources for Semantic Mediation, 21st International Conference on Conceptual Modeling (ER). 2002.
[14] Martone, M.E., Gupta, A., Wong, M., et al, A Cell-Centered Database for Electron Tomographie Data. Journal of Structural Biology 2002;138:145–155. http://ncmir.ucsd.edu/CCDB/
[15] Artale, A., Franconi, E., Guarino, N., et al. Part-Whole Relations in Object-Centered Systems: An Overview. Data and Knowledge Engineering. 1996;20:347–383.
[16] Mitra, P., Wiederhold, G., Kersten, M.L., A Graph-Oriented Model for Articulation of Ontology Interdependencies, Proceedings of the International Conference on Extending Database Technology (EDBT). 2000:86–100.
[17] Horrocks, I.R. Using an Expressive Description Logic: FaCT or Fiction? In: Cohn A.G., Schubert L., Shapiro S.C., eds. KR’98: Principles of Knowledge Representation and Reasoning. San Francisco: Morgan Kaufmann; 1998:636–645.
[18] Ludäscher, B., Gupta, A., Martone, M.E. Model-Based Mediation with Domain Maps. In: Proceedings of the 17th International Conference on Data Engineering (ICDE). New York: IEEE Computer Society; 2001.
[19] Calvanese, D., Giacomo, G.D., Lenzerini, M., et al, Description Logic Framework for Information Integration, Principles of Knowledge Representation and Reasoning. 1998:2–13.
[20] Ludäscher, B., Gupta, A., Martone, M.E. Model-Based Mediation with Domain Maps. In: Proceedings of the 17th International Conference on Data Engineering (ICDE). New York: IEEE Computer Society; 2001:81–90.
[21] July Kifer, M., Lausen, G., Wu., J. Logical Foundations of Object-Oriented and Frame-Based Languages. Journal of the ACM. 1995;42(no. 4):741–843.
[22] FLORA homepage, http://www.cs.sunysb.edu/∼sbprolog/flora/.
[23] FLORID homepage, http://www.informatik.uni-freiberg.de/∼dbis/florid/.
[24] Ludäscher, B., Gupta, A., Martone, M.E. Model-Based Information Integration in a Neuroscience Mediator System. In: Proceedings of the 26th International Conference on Very Large Data Bases (VLDB). San Francisco: Morgan Kaufmann; 2000:639–642.
[25] Valencia, Spain Himmeröder, R., Lausen, G., Ludäscher, B., et al, FLORID: A DOOD-System for Querying the Web, Demonstration Session at EDBT. 1998.
[26] Ludäscher, B., Himmeröder, R., Lausen, G., et al. Managing Semistructured Data with FLORID: A Deductive Object-Oriented Perspective. Information Systems. 1998;23(no. 8):589–613.
[27] Lecture Notes in Computer Science 1472 Lausen, G., Ludäscher, B., May, W., On Active Deductive Databases: The Statelog Approach. Springer. Freitag B., Decker H., Kifer M., et al, eds. Transactions and Change in Logic Databases. 1998:69–106.
[28] July Kifer, M., Lausen, G., Wu., J. Logical Foundations of Object-Oriented and Frame-Based Languages. journal of the ACM. 1995;42(no. 4):741–843.
[29] Lecture Notes in Computer Science 2287 Qian, X., Ludäscher, B., Martone, M.E., et al, Navigating Virtual Information Sources with Know-ME, EDBT. 2002:739–741.
[30] Ludäscher, B., Gupta, A., Martone, M.E. Model-Based Information Integration in a Neuroscience Mediator System. In: Proceedings of the 26th International Conference on Very Large Data Bases (VLDB). San Francisco: Morgan Kaufmann; 2000:639–642.
[31] Yang, G., Kifer, M., FLORA: Implementing an Efficient DOOD System Using a Tabling Logic Engine, Sixth International Conference on Rules and Objects in Databases (DOOD). 2002.
[32] technical report Draxler, C. A Powerful Prolog to SQL Compiler. München, Germany: Centre for Information and Language Processing, Ludwigs- Maximillians-Universität München; 1992.
[33] Ludäscher, B. Mediator Query Processing with Prolog Technology. In: technical note, BIRN-DI-TN-2002-01. Biomedical Informatics Research Network; 2002.
[34] Sagonas, K.F., Swift, T., Warren, D.S., XSB as an Efficient Deductive Database Engine, Proceedings of the ACM International Conference on Management of Data (SIGMOD). 1994:442–453.
[35] Zaslavsky, I. A New Technology for Interactive Online Mapping. Cartographic Perspectives. 2000;37:65–77.
[36] Paxinos, G., Watson, C. The Rat Brain in Stereotaxic Coordinates. San Diego: Academic Press; 1998.
[37] Lecture notes in Computer Science 2405. Springer Gupta, A., Ludäscher, B., Martone, M.E., et al, A System for Managing Alternate Models in Model-Based Mediation, Advances in Databases, 19th British National Conference on Databases (BNCOD 19). 2002.
[38] Martone, M.E., Gupta, A., Ludäscher, B., et al. Federation of Brain Data through Knowledge-Guided Mediation. In: Kötter R., ed. Neuroscience Databases: A Practical Guide. Boston: Kluwer Academic Publishers; 2002:275–292.
[39] (Extended Abstract) Garcia-Molina, H., Papakonstantinou, Y., Quass, D., et al, The TSIMMIS Approach to Mediation: Data Models and Languages, Next Generation Information Technologies and Systems. 1995.
[40] Knoblock, C.A., Minton, S., Ambite, J.L., et al, Modeling Web Sources for Information Integration, Proceedings of the Fifteenth National Conference on Artificial Intelligence. 1998.
[41] Levy, A.Y., Rajaraman, A., Ordille, J.J., Querying Heterogeneous Information Sources Using Source Descriptions, Proceedings of the International Conference on Very Large Data Bases (VLDB). 1996:251–262.
[42] Haas, L.M., Kossmann, D., Wimmers, E.L., et al, Optimizing Queries Across Diverse Data Sources, Proceedings of the International Conference on Very Large Data Bases (VLDB). 1997:276–285.
[43] Baru, C., Gupta, A., Ludäscher, B., et al. XML-Based Information Mediation with MIX. In: Proceedings of the ACM SIGMOD International Conference on Management of Data (SIGMOD 1999). Philadelphia: ACM Press; 1999:597–599.
[44] Palopoli, L., Terracina, G., Ursino, D., The System DIKE: Towards the Semi-Automatic Synthesis of Cooperative Information Systems and Data Warehouses, Proceedings of the ADBIS-DASFAA Symposium. 2000:108–117.
[45] Beneventano, D., Bergamaschi, S., Extensional Knowledge for Semantic Query Optimization in a Mediator-Based System, International Workshop on Foundations of Models for Information Integration (FMII-2001). 2001.
[46] Bergamaschi, S., Castano, S., Vincini, M. Semantic Integration of Semistructured and Structured Data Sources. SIGMOD Record. 1999;28(no. 1):54–59.
[47] Calvanese, D., Castano, S., Guerra, F., et al, Towards a Comprehensive Methodological Framework for Semantic Integration of Heterogeneous Data Sources, Eighth International Workshop on Knowledge Representation Meets Databases (KRDB). 2001.
[48] Peim, M., Franconi, E., Paton, N., et al, Query Processing with Description Logic Ontologies Over Object-Wrapped Databases, Proceedings of the International Conference on Scientific and Statistical Database Management (SSDBM). 2002.
[49] Goble, C., Stevens, R., Ng, G., et al. Transparent Access to Multiple Bioinformatics Information Sources. IBM Systems Journal. 2001;40(no. 2):534–551.
[50] Gupta, A., Ludäscher, B., Martone, M.E. Knowledge-Based Integration of Neuroscience Data Sources. In: Proceedings of the Twelfth International Conference on Scientific and Statistical Database Management (SSDBM). IEEE Computer Society; 2000:39–52.
[51] Horrocks, I., DAML+OIL: A Reason-Able Web Ontology Language, Proceedings of the International Conference on Extending Database Technology (EDBT). 2002:2–13.
1Such simple mediation scenarios often pose very difficult technical challenges (e.g., query processing in the presence of limited source capabilities) [1, 2].
2Information about the two laboratories SYNAPSE and NCMIR is respectively available at http://synapses.bu.edu and http://www-ncmir.ucsd.edu.
3The w3c working group XML Schema (http://www.w3.org/XML/Schema) and similar efforts like RELAX NG (http://www.oasis-open.org/committees/relax-ngl) play an intermediate role between purely structure-based models (DTDs) and richer semantic models with constraint mechanisms.
4Most formal approaches (e.g., those based on description logic) consider binary relationships only.
5For example, ONT4, ONT5 in Figure 12.4 define virtual relations such as transitive closure over the base relations.
6By standards of meronyms, there are different kinds of the has relation, including component-object has (co), portion-mass has (pm), member-collection has (mc), stuff-object has (so), and place-area has (pa) [15].
7The Unified Medical Language System (UMLS) available at http://www.nlm.nih.gov/researcb/umls/ is, strictly speaking, a metathesaurus, or a semi-formal ontology with a limited set of pre-defined relationships such as broader-term/narrower-term.
8See http://www.geneontology.org/process.ontology for information about the Biological Process from the Gene Ontology Consortium.
9At the cost of loss of efficiency, the restriction no recursion through the Web could be lifted.
10This is similar to the fact that the same query can have many different syntactic representations. In general, equivalence of first-order (or SQL) queries is not decidable.
11Thus, C and D can be viewed as unary predicates.
12RDF(S) has such a notion called subproperty; see http://www.w3.org/TR/rdf-scbema/.
13Strictly speaking, the isa does not have to be a hierarchy but can be any directed acyclic graph.
14Because concepts are implemented as F-logic classes, this avoids terminological cycles.
15This is reversed from the usual convention used in the rest of the chapter to match this DM notation.
16Here, NProt stands for nuclear protein.
17For example, orchestra has_a musician and musician has_a arm, but not orchestra has_a arm.
18See Lausen et al., Section 6 [27] for a formalization of hierarchical processes using nested Kripke structures.
19The National Center for Microscopy and Imaging is a research facility specialized in the development of technologies for improving the understanding of biological structure and function relationships spanning the dimensional range from 5nm3 to 50μm3 (http://www.ncmir.ucsd.edu).