Complex Query Formulation Over Diverse Information Sources in TAMBIS
Robert Stevens, Carole Goble, Norman W. Paton, Sean Bechhofer, Gary Ng, Patricia Baker and Andy Brass
Molecular biology is a data-rich discipline that has produced a vast quantity of sequence and other data. Most of the resulting data sets are held in independently developed databanks and are acted upon by separate analysis tools. These information sources and tools are autonomous, distributed, and have differing call interfaces. As such, they manifest classical syntactic and semantic heterogeneity problems [1].
Many bioinformatics tasks are supported by individual sources. However, biologists increasingly wish to ask complex questions that span a range of the available sources [2]. This places barriers between a biologist and the task to be accomplished; the biologist has to know what sources to use, the locations of the sources, how to use the sources (both syntactically and their semantics), and how to transfer data between the sources.
This chapter presents an approach to solving these problems called Transparent Access to Multiple Bioinformatics Information Sources (TAMBIS) [3]. This chapter reports on the first version of the TAMBIS system, which was developed between 1996 and 2000. A second version extends and develops this first version, addressing some of the problems recognized in the approach. This new version is introduced in Section 7.5. The TAMBIS approach attempts to avoid the pitfalls described previously by using an ontology of molecular biology and bioinformatics to manage the presentation and usage of the sources. An ontology is a description of the concepts, and the relationships between those concepts, within a domain.
![]() to provide a homogenizing layer over the numerous databases and analysis tools
to provide a homogenizing layer over the numerous databases and analysis tools
![]() to manage the heterogeneities between the data sources
to manage the heterogeneities between the data sources
![]() to provide a common, consistent query-forming user interface that allows queries across sources to be precisely expressed and progressively refined
to provide a common, consistent query-forming user interface that allows queries across sources to be precisely expressed and progressively refined
This ontology is the backbone of the TAMBIS system; it is what the user interacts with to form questions. It allows the same style of query and terms to be used across diverse resources, and it also manages the answering of the query itself.
A concept is a description of a set of instances, so a concept or description can also be viewed as a query. The TAMBIS system is used for retrieving instances described by concepts in the model. This contrasts with queries phrased in terms of the structures used to store the data, as are used in conventional database query environments. This approach allows a biologist to ask complex questions that access and combine data from different sources. However, in TAMBIS, the user does not have to choose the sources, identify the location of the sources, express requests in the language of the source, or transfer data items between sources.
Figure 7.1 shows how a query is constructed and processed through the TAMBIS system. The steps in processing a TAMBIS query are as follows:
1. A query is formulated in terms of the concepts and relationships in the ontology using the visual Conceptual Query Formulation Interface. This interface allows the ontology to be browsed by users and supports the construction of complex concept descriptions that serve as queries. The output of the query formulation process is a source independent conceptual query. The query formulation interface makes extensive use of the TAMBIS Ontology Server, which not only stores the ontology but supports various reasoning services over the ontology. These reasoning services serve, for example, to ensure that queries constructed using the query formulation interface are biologically meaningful with respect to the TAMBIS ontology.
2. Given a query, TAMBIS must identify the sources that can be used to answer the query and construct valid and efficient plans for evaluating it given the facilities provided by the relevant sources. The source selection and query planning process makes extensive use of the Sources and Services Model (SSM), which associates concepts and relationships from the Ontology with the services provided by the sources. The output of the source selection and query planning process is a source dependent query plan that describes the sources to be used and the order in which calls should be made to the sources.
3. The query plan execution process takes the plan provided by the planner and executes that plan over the wrapped sources to yield an answer to the query. Sources are wrapped so they can be accessed in a syntactically consistent manner. In version one of TAMBIS, each source is represented as a collection of function calls, which are evaluated by the collection programming language (CPL) [4]. The sources used in TAMBIS 1.0 were Swiss-Prot, ENZYME, CATH (Classes, Architecture, Topology, Homology), Basic Local Alignment Search Tool (BLAST), and PROSITE.
The remainder of this Chapter is organized as follows. Section 7.1 gives a brief overview of the TAMBIS ontology, describing its scope and the language in which it is implemented. Section 7.2 describes how users interact with TAMBIS, in particular how the ontology is explored and how queries are constructed using the interface from Section 7.2. Section 7.3 describes how queries constructed using the interface from Section 7.2 are evaluated over the individual sources. Section 7.4 describes work in several areas related to TAMBIS and describes how TAMBIS compares to alternative or complementary proposals. Section 7.5 considers issue relating to query construction and source integration raised by experience in TAMBIS and how these are addressed in TAMBIS 2.0.
7.1 THE ONTOLOGY
An ontology is a description of the concepts and their relationships within a domain. An ontology is a mechanism by which knowledge about a domain can be captured in computational form and shared within a community [5]. The TAMBIS ontology describes both molecular biology and bioinformatics tasks.
A concept represents a class of individuals within a domain. Concepts such as Protein and Nucleic acid are part of the world of molecular biology. An Accession number, which acts as a unique identifier for an entry in an information source, lies outside this domain but is essential for describing bioinformatics tasks in molecular biology. The TAMBIS ontology contains only concepts and the relationships between those concepts. Individuals that are members of concept classes (P21598 is an individual of the class Accession number) do not appear in the TAMBIS ontology. Such individuals are contained within the external resources over which TAMBIS answers queries.
The TAMBIS ontology has been designed to cover the standard range of bioinformatics retrieval and analysis tasks [2]. This means that a broad range of biology has been described. The model is, however, currently quite shallow; although the detail present is sufficient to allow descriptions of most retrieval tasks supportable using the integrated bioinformatics sources. In addition, precision can arise from the ability to combine concepts to create more specialized concepts (see Section 7.2.2).
The model is centered upon the biopolymers Protein and Nucleic acid and their children, such as Enzyme, DNA, and RNA. Biological functions and processes are also present, so it is possible to describe, for example, the kinds of reactions that are catalyzed by an enzyme. Many tasks in bioinformatics involve comparing or identifying patterns in sequences. As a result, sequence components such as protein motifs and structure classifications are described. For example, a motif is a pattern within a sequence that is generally associated with some biological function. The ontology thus supports the description of motifs and various different kinds of motifs. Such descriptions are facilitated by the presence of a rich collection of relationships between concepts in the ontology. These basic concepts are present in the is a hierarchy. Other relationships add richness to the model, so that a wide range of biological features can be described. For example, Motif (and its children) can be components (parts of) Protein or Nucleic acid. Other relationships capture associations to functions, processes, sub-cellular locations, similarities, and labels such as species name, gene names, protein names, and accession numbers. The model is described in more detail in an article in Bioinformatics [6] and can be browsed via an applet on the TAMBIS Web site.
The TAMBIS ontology is expressed in a description logic (DL) [7], a type of knowledge representation language for describing ontologies [8]. DLs are considered an important formalism for giving a logical underpinning to knowledge representation systems, but they also provide practical reasoning facilities for inferring properties of and relationships between concepts [9]. TAMBIS makes extensive use of these reasoning services.
As well as the traditional isa relationships (e.g., a Motif isa SequenceComponent), there are partitive (describing parts), locative (describing location), and nominative (describing names or labels) relationships. This means that the TAMBIS ontology can describe relationships such as: “Motifs are parts of proteins” and “Organelles are located inside cells.” The ontology initially holds only asserted concepts, but these can be combined dynamically via relationships to form new, compositional concepts. These compositional concepts are automatically classified using the reasoning services of the ontology. Such compositional concepts can be made in a post-coordinated manner: That is, the ontology is not a static artifact; users can interact with the ontology to build new concepts, composed of those already in the ontology, and have them checked for consistency and placed at the correct position in the ontology’s lattice of concepts. For example, Motif can be combined with Protein using the relationship isComponentOf to form a new concept Protein motif, which is placed as a kind of Motif.
The ontology is a dynamic model in that what is present in the model is the description of potential concepts that can be formed in the domain of molecular biology and bioinformatics. As these new, compositional concepts are described, they are placed automatically within the lattice of existing concepts by the DL reasoning services. For example, the compositional concept Protein motif (see above) is automatically classified as a kind of Motif. This new concept is then available to be re-used in further compositional concepts. Most of the other biological ontologies are static; the TAMBIS ontology is dynamic, built around a collection of concept descriptions and constraints on how they can be composed.
The TAMBIS ontology is described using the DL called Galen Representation and Integration Language (GRAIL) [10]. In GRAIL, a new concept can be defined as follows:
![]()
where each ri is a role name and each fi a filler concept. Each rifi pair is also known as a criterion. A role is a property of a concept, and the filler of a role is the name or description of the concept that can play the given role. For example, Motif which isComponentOf Protein is a description of a protein motif. Motif and Protein are names of existing concepts, which are acting here as the base concept and a role filler respectively. The construct isComponentOf Protein is the criterion of Motif in this case.
Description logic ontologies are organized within a subsumption lattice, which captures the isa relationship between two concepts. The fact that one concept is a kind of another can either be asserted as part of the model, or inferred by the reasoning system on the basis of the concept descriptions. Figure 7.2 illustrates both forms of subsumption relationship. For example, Motif has been asserted to be a kind of SequenceComponent, and PhosphorylationSite has been asserted to be a kind of Motif. By contrast, with the asserted hierarchy, the notion of a Motif that can be found within a protein (Motif which isComponentOf Protein) is inferred to be a kind of Motif, as are the other concepts in the three boxes on the bottom in Figure 7.2. In these cases, the criteria describing the concept are used to infer the classification of these concepts. Wherever C2 is subsumed by C1, every instance of C2 is guaranteed to be an instance of C1 (e.g., every Motif is a SequenceComponent, and every Motif which isComponentOf Protein is a Motif).

FIGURE 7.2 Example of subsumption relationship within the ontology. The concepts that have been inserted into the lattice are shaded in the three boxes at the top of the figure. The locations of the unshaded concepts in the lattice have been inferred.
In spite of its inexpressiveness compared with some other DLs [11], the GRAIL representation has a useful property in its ability to describe constraints about when relationships are allowed to be formed. For example, it is true that a Motif is a component of a Biopolymer, but not all motifs are components of all biopolymers. For example, a PhosphorylationSite can be a component of a Protein, but not a component of a Nucleic acid, both of which are Biopolymers. The constraint mechanism allows the TAMBIS model to capture this distinction and thus only allow the description of concepts that are described as being biologically meaningful in terms of the model from which they are built. This allows general queries, such as “find all protein motifs,” to be expressed as well as specific queries such as “find phosphorylation motifs upon this protein.”
The TAMBIS ontology is supplied as a software component that acts as a server. Other components can ask questions of the knowledge in the ontology component. It is the backbone of the architecture, and other components either directly or indirectly use the ontology. These other components ask questions such as: “is this a concept;” “what are the parents, children, or siblings of this concept;” “which relationships are held by this concept;” and “what is the natural language version of this concept.”
7.2 THE USER INTERFACE
This section describes the user interface to TAMBIS. The interface supports users in carrying out two principal tasks: exploring the ontology and constructing queries, which are described in Sections 7.2.1 and 7.2.2, respectively.
7.2.1 Exploring the Ontology
Although the full TAMBIS ontology contains approximately 1800 concepts, the version of the ontology used in the online system for querying contains approximately 250 concepts. This model concentrates on proteins and enzymes; it describes features such as functions, processes, motifs, and structure. In this and following sections, examples are based on this smaller ontology.
The main window of the TAMBIS system is shown in Figure 7.3. The main window is used to launch exploration or query building tasks. A concept name is either typed into the find field directly or obtained from the list of Bookmarks. This concept can be used either as the starting point for model exploration or query building by selecting New query or Explore. If Explore is selected in Figure 7.3, the explorer window depicted in Figure 7.4 is launched.
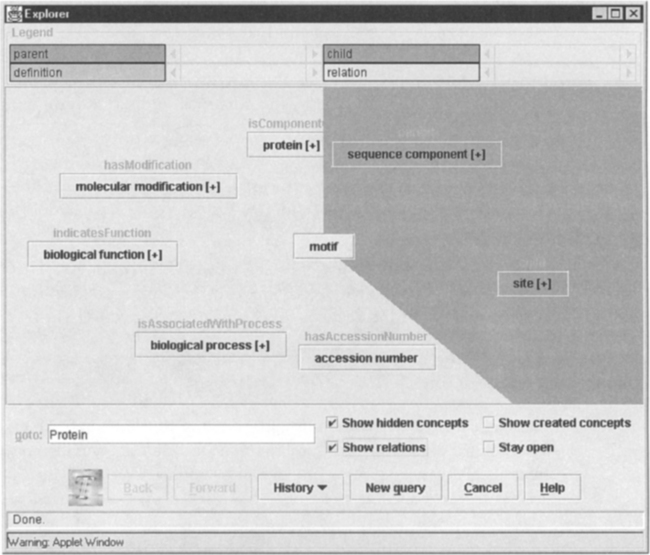
FIGURE 7.4 The explorer window showing motif with all types of relations it has with other concepts.
The window in Figure 7.4 shows the basic concept description facilities of the model browser. Concepts are shown as buttons; the buttons usually have a title that describes the relationship to the central or focus concept, which has no title itself. The button color also indicates the relationship the button has to the central concept, although this is not evident in the monochrome screenshot.
Figure 7.4 shows all the relationships of motif. The parent and children concepts are sequence component and site, respectively. The relationships other than is-a-kind-of are shown in the lighter area of the figure. The name of the relationship appears as the button title, and the name of the concept to which the relationship links is the button label. For example, a relationship button title is hasAccessionNumber and a concept button label is accession number. Table 7.1 shows these relationships for motif. The user can explore the is-a-kind-of hierarchy or the other relationships by clicking on the buttons representing the concepts to which motif is related.
TABLE 7.1
The relationships from motif to other concepts in the TAMBIS model.
| Relationship | Concept |
| has Accession Number | accession number |
| is Component of | protein |
| indicates Function | biological function |
| is Associated With Process | biological process |
| has Modification | molecular modification |
The explorer uses a pie-chart view of the ontology, with different sectors showing the parents, children, definitions, and other relationships. In the TAMBIS ontology, some concepts have a large number of members in one sector, far more than can be shown at any one time. Rather than cramping the view of related concepts, the sectors are scrollable, allowing controlled viewing of the ontology’s contents.
Clicking on a concept button that is not the focus causes that button to become the new focus. Thus, a user can move up and down the taxonomy and across the taxonomies by following other relationships. Larger jumps may be made within the model by using a go to function.
7.2.2 Constructing Queries
Queries in TAMBIS are essentially concept descriptions. Thus, the task of query formulation involves the user in constructing a concept that describes the information of interest. An example query is illustrated in Figure 7.5, which is a screen shot of the query builder window containing a request for the motifs that are components of guppy proteins. The equivalent GRAIL concept is:
![]()
As its name indicates, the query builder window is used for building descriptions of biological concepts that act as queries. One of the buttons along the bottom of the window, Submit, is used to ask TAMBIS to process the query and collect the results. Part of a results page for this query is given in Figure 7.6. The results shown in this figure are the values for the base concept of the query (i.e., the properties of the motifs that are components of guppy proteins). This set of results contains only the base concept (Motif); other concepts may be included in the results and the relationships maintained between the different instances via the query builder. The pop-up menu on a concept button contains an option include in results. Selecting this option causes the concept button to be highlighted in the query builder.
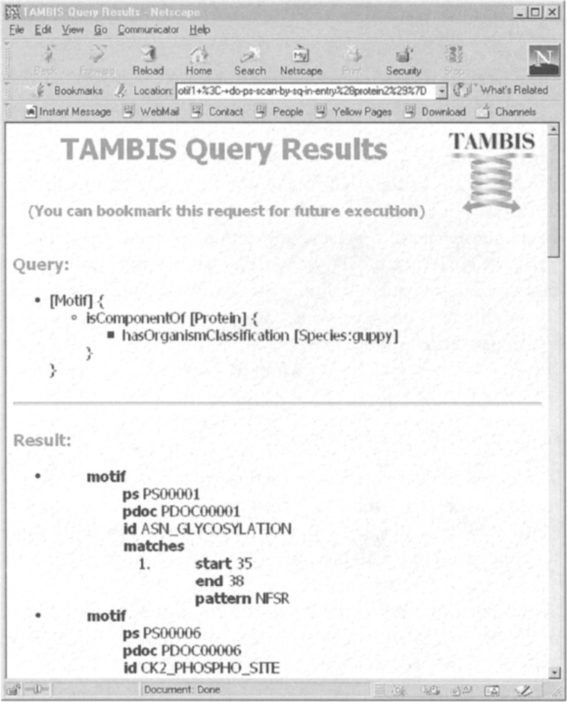
FIGURE 7.6 Part of the results page that fulfills the description shown in Figure 7.5.
Given that a query is of the form:
![]()
where each ri is a role name and each fi a filler concept, the query builder essentially supports:
1. The specialization or generalization of the base or filler concepts
2. The addition or removal of criteria associated with a composite concept
This incremental concept construction and modification is possible because of the dynamic model supported by the ontology. In fact, the query interface is driven directly from the model, and the reasoning services are used extensively during query construction to present appropriate options to users and for validating the concepts constructed. The knowledge held in the ontology is used to guide the user through the query building process by offering only appropriate possibilities for modifying a query at each stage [12]. As new concepts are formed and classified, new criteria become available and others are lost as potential additions to the growing concept. Support for the previous query construction operations is illustrated in the following subsections.
Replacing Part of a Query
The query builder can be used either to construct a query from scratch or for modifying previous or bookmarked queries. One way of modifying the query is to replace the concept motif with one that is more specific. An example is replacing the concept motif with the more specialized concept phosphorylation site.
Figure 7.7 shows a menu associated with concepts in the query builder. Selecting replace with-a-kind-of-this causes a new window to appear, as shown in Figure 7.8. This window is the replacer window, which allows a version of the explorer to be used to identify a concept that can be used in the query in place of motif.

FIGURE 7.7 A query builder window showing the pop-up menu invoked by clicking on the topic concept motif.
When launched, the replacer is focused on the motif concept, on which the query builder had focus. Moving down the hierarchy, through site to modified site yields the window shown in Figure 7.9. Selecting phosphorylation site and pressing replace it updates the query in the query builder so that the query has the same structure as that in Figure 7.5, but with motif replaced with phosphorylation site (Figure 7.10). The replacer limits the user to the is a hierarchy during replacement. This helps ensure that only valid concepts are created.
Restricting a Concept
When one concept is joined to another in the query builder with a relationship other than is-a-kind-of, the description of the original concept is restricted. In the example query, motifs are restricted to those that occur in proteins, rather than Motifs that can occur in other kinds of molecules. This restriction was added to motif using the restrict by a relationship option illustrated in Figure 7.7. The type of motif to be retrieved can be further restricted by adding another concept to the description of motif. For example, selecting the restrict by a relationship option leads to the user being offered the restrict window shown in Figure 7.11. If the user then selects the hasModification post translational modification check box and accept, the query in the query builder is replaced with that in Figure 7.12. The query is now “retrieve all motifs that bring about post translational modifications in guppy proteins.”
Nonsensical Questions
The TAMBIS model only allows biologically sensible questions to be constructed. By only allowing is-a-kind-of relationships to he seen in the replacer, the tendency is to have only biologically sensible queries constructed. It is, however, possible to replace a valid concept with one that is biologically nonsense. However, the query builder detects this by consulting the ontology and informs the user of the error.
For example, in the previously modified query it would be possible to replace the concept protein with the concept nucleic acid. However, if this replacement is made, TAMBIS notices that in the ontology nucleic acids cannot have phosphorylation sites and changes the color of the offending concept button (nucleic acid in this case) to yellow, indicating that the query is not consistent with the constraints in the ontology.
7.2.3 The Role of Reasoning in Query Formulation
GRAIL, like other DL implementations, provides a classification or reasoning service, which allows the organization of concept descriptions into subsumption (isa) hierarchies. In the case of DLs, this is most interesting when applied to composite descriptions. In standard taxonomies, the position of each concept is explicitly stated by the modeler. Within TAMBIS, through the use of the ontology server, the position of composite concept descriptions can be determined by the reasoner. This is of particular importance when new, previously unseen, descriptions are introduced into the model—particularly when a user forms a new concept to ask a query.
The basic classification hierarchy can be used to navigate through the existing descriptions in the model (e.g., using the explorer). More interesting, however, is TAMBIS’s ability to support the formation of new, composite query expressions (through the use of the query builder).
TAMBIS uses a constraint mechanism known as sanctioning to drive the query builder user interface [10]. Information included in the ontology specifies the compositions that may be formed, and this in turn determines the specialization options that may be applied to a query. This type of constraint mechanism is peculiar to DLs, as such constraints naturally form part of many frame-based knowledge representation languages. These constraints are, however, important in describing what concepts are allowed to be formed within the ontology.
For example, the concept Motif may be restricted or specialized through a number of relationships including hasOrganismClassification or indicatesFunction. For each of these relationships, the allowable values are constrained by the values of the sanctions in the model. For example, hasOrganismClassification can only be filled with the concept kingdom (or one of its subclasses).
It would be an onerous task to specify explicitly the potential values for any combination, so to minimize the information required, sanctions are inherited down the classification hierarchy in the model. Thus, the sanctioning information can be added sparsely. As a query is gradually built up, its position in the classification hierarchy will change, leading to changes in the restriction options offered. The reasoner is key to this process because it is used to determine the appropriate position of a query description and thus, the potential restrictions. As the query is constructed, the interface communicates with the ontology server, updating the restrictions offered to the user. The constraints or sanctions can also be viewed through the explorer; the relationships shown are exactly those that can be used for specialization or restriction of the concept in a query.
7.3 THE QUERY PROCESSOR
The query processor converts a source independent declarative GRAIL query into a source specific execution plan expressed in CPL [4]. CPL allows the concise expression of retrieval requests over collections of data, with data types for representing arbitrarily nested sets, bags, lists, records, and variants. The principal components of the query processor are the wrappers, the sources and services model (SSM), and the planner.
7.3.1 The Sources and Services Model
The SSM stores the relationships between the concepts and roles in the ontology and the functions used to wrap sources in CPL. In the SSM, the ontology is used to index the CPL functions used to evaluate queries written in terms of the ontology. The SSM contains descriptions of three broad categories of information: iterators that retrieve instances of concepts from sources, role evaluators that retrieve or compute values for the roles of instances, and filters that are used to discard instances not relevant to the query.
Each such description of a CPL function in the SSM includes its name, the types of its arguments, the type of its result, some information on the cost of computing the function, and the source accessed by the function.
There are seven categories of mapping information supported within the SSM, which are described in detail in a paper of the Proceedings of 11th International Conference on Scientific and Statistical Data Management [13]. Four of these categories are described here to illustrate how the query processor works:
1. Iteration: Iteration allows the instances of a concept to be retrieved from a source. For example, the fact that the instances or individuals of Protein can be obtained from Swiss-Prot is represented by associating the concept Protein with the function get-all-sp-entries, which has no input arguments and which returns results of type protein_record. Given a query in which the instances of protein are required, this SSM entry could be used to retrieve proteins from Swiss-Prot using a function call such as:
![]()
If an alternative, more specialized source of protein information is available, for example, from a database of enzymes (any protein that acts as a catalyst is an enzyme), then an additional SSM entry can be created to indicate this. In fact, there is a source called ENZYME that stores descriptions of enzymes, and thus, there is an SSM entry associating the concept Protein which hasFunction catalysis with a function get-all-enzyme-entries that supports iteration over the entries in the ENZYME database. During query processing, the planner uses the most specialized source of information available to answer a query. If there are several sources of the same information (e.g., if there is more than one protein source), this must be handled within the wrappers. This restriction within the first version of TAMBIS is to be relieved in future versions of TAMBIS (see Section 7.5).
2. Roles: Roles allow the evaluation of a role in an instance to obtain a value for its filler. For example, it is possible to obtain the AccessionNumber of a protein given the Protein. This is represented in the SSM by the association of the concept Protein which hasAccessionNumber AccessionNumber with the function get-ac-from-sp-entry, which takes as argument a value of type protein_record and returns a value of type accession_number. This does not itself directly access a source, but rather it accesses a data structure retrieved from a source by some other function (such as get-all-sp-entries described previously). This SSM entry could be used to retrieve the accession number from a Swiss-Prot entry using a function call such as:
![]()
where p is a variable previously bound to a protein_record.
3. Mapped Roles: Mapped roles are roles in which the concept provided as the role filler can be used to select instances of the base concept from a source. For example, instances of the concept Protein which hasOrganismClassification Species: guppy can be retrieved from Swiss-Prot by retrieving entries with guppy in their organism species field. This SSM entry could be used to retrieve Swiss-Prot entries using a function call such as:
![]()
4. Filters: When instances of a concept have been retrieved, for example, by iteration, other criteria in the query may be used to discard some of the instances. For example, given an instance of Protein in the query Protein which hasFunction Hydrolase, the instance of Protein must be checked to see if it hasFunction Hydrolase. The relevant SSM entry could be used to generate code that tests a Swiss-Prot record for the function hydrolase using a function call such as:
![]()
where p is a variable previously bound to a protein_record.
The filters entries in the SSM are used to select values with the required characteristics at the client (i.e., values are retrieved from sources and then checked to see if they meet the needs of the query). In general, it is desirable to have the sources retrieve only values that are relevant. Mapped roles provide one way of sending filters to the sources to be applied as early as possible in the retrieval process. Unfortunately, at the time of writing, many sources did not offer query interfaces that allowed all filtering to be carried out early in the query process. This left much client-side filtering to take place.
7.3.2 The Query Planner
GRAIL queries are declarative, in that the meaning of a query is not dependent on the order of evaluation of its components. As a result, the TAMBIS system, and not the user, must take responsibility for identifying an efficient evaluation order for the components of a GRAIL query. This section describes how GRAIL queries are represented internally for the purposes of optimization and how this internal representation is generated.
GRAIL queries are intrinsically nested structures. The query internal form (QIF) used in TAMBIS can be seen as an un-nested representation of the original GRAIL query. This representation has been developed to allow easier reordering of the components of a query in the planner. The QIF is a list of query components, an example of which is given in Figure 7.13 for the running example GRAIL query:
![]()
The query is represented by two query components, one representing the Motif and the other representing the Protein. Each of the components stores the name of the base concept, a list of the criteria from the query, the name of the CPL variable used to hold values retrieved from sources, and details of the technique identified by the planner for retrieving instances of the concept and of the criteria used during retrieval. The values for theTechnique and theFetchCriterion are identified during planning. Generation of the QIF from a GRAIL query is straightforward and is carried out in a single pass over the query.
Given the QIF for a query, a search algorithm seeks to identify efficient ways to evaluate the query given the functions available in the SSM. The search algorithm exploits the augmentation heuristic [14], which was selected because it is straightforward to implement and provides a reasonable tradeoff between cost of optimization and quality of plan generated. The algorithm is given in Figure 7.14. The basic strategy is to generate a plan as an ordered list of query components in which the first component in the list is predicted to be the least costly component to evaluate from scratch, and the subsequent components are the least costly to evaluate given what has previously been evaluated.
The optimization algorithm in Figure 7.14 depends heavily on the definition of the findBest function. This function, given a query component, considers a variety of ways in which instances of the component can be retrieved from sources. Thus findBest considers the alternative ways of implementing the components of a QIF onto CPL functions, using the entries in the SSM.
For example, the CPL generated for the example query is:

This query contains two query components, one for Protein and the other for Motif, as illustrated in Figure 7.13. The query component for Protein is chosen for evaluation first, and the SSM entry used to obtain instances of Protein is the role that is the inverse of hasOrganismClassification. Thus, the query processor accesses the ontology to find the inverse of hasOrganismClassification, which is the role hasProteins on Species. This role has a roles entry in the SSM, which is associated with the function get-sp-entry-by-os. This function, given the name of a Species, consults Swiss-Prot to find the proteins from the species. The second query component is evaluated in a similar manner, using the inverse of the role isComponentOf.
The output from the planner is a QIF annotated with details of how to retrieve its components. Generating the corresponding CPL program involves a single pass through the QIF. For each QIF component, the code generator writes out the CPL functions identified by the planner and iterates over the component’s other criteria, writing out function calls associated with roles and filters as required.
7.3.3 The Wrappers
The distribution and heterogeneity within bioinformatics resources means that many applications need to employ wrappers. Wrappers include external resources into a system that enable the resource to adopt the same operating paradigms as the host system, as well as transform the resource to common syntactic and semantic conventions. Many applications perform this wrapping on an ad hoc basis, using the resources available within many programming languages. Kleisli (presented in Chapter 6) is one of the few systems to offer wrapper services together with a query language that is flexible enough to cope with bioinformatics resources.
The output from the TAMBIS system is a query plan written in CPL using a modified version of the BioKleisli library of biological database wrappers [15]. An example CPL query, which “retrieves all motifs in guppy proteins,” is as follows:

In the query, the part before the | is the projection expression, which, in this case, indicates that only the motifs m are of interest. The two function calls in the body of the query to the right of the | are generators, which retrieve values from distinct, wrapped sources. The first line in the query body indicates that the new variable p is to be bound to each of the values that result from the evaluation of the function get-sp-entry-by-os with the parameter guppy. The function name can be read as get Swiss-Prot entry by organism species, although this is just a name—the structure of the name is not significant in itself. The second function call binds the variable m to each of the motifs of the proteins bound to p. The function name can be read as scan the prosite database for motifs in the given protein record.
The CPL system is supplied with function libraries that provide access to a range of bioinformatics sources of different types (e.g., databases, analysis tools [15]). TAMBIS uses these libraries and a number developed to provide a function-based view of the sources. The public release of TAMBIS 1.0 accessed five sources and used a total of approximately 300 CPL functions.
CPL can be seen as providing syntactically consistent, but not source transparent, access to the sources, and thus, CPL can be viewed as a wrapping mechanism tightly coupled with convenient language facilities for accumulating and transmitting results from different sources.
Remarks on Handling Syntactic and Semantic Heterogeneity
The heterogeneity in the bioinformatics resources is handled within this wrapper layer and the SSM. The wrapper layer irons out much of the structural or syntactic heterogeneity, providing a consistent call interface in terms of level of abstraction and services to each of the resources. For example, all CPL functions return sets of data, regardless of the number of instances returned. This means only one operator ever needs to be used to manipulate the results of a query. Any heterogeneity in encoding, such as representation of amino acid sequences, can also be dealt with at this level.
The wrapper layer also gives an opportunity for standardization of naming conventions for services available in the resources, though this is of no consequence to users, except that they may find the CPL query plan useful as a quality check on the task TAMBIS is performing.
The SSM affords the main opportunity for the reconciliation of semantic heterogeneity. The ontology gives the user a global schema against which to form queries. The SSM allows terms seen in this global schema to be mapped to the values used in the various resources. For instance, the concept PhosphorylationSite corresponds to the motif entry PS00001 in the PROSITE databank. Similarly, the concept Kinase maps to node 2.7.*.* in the ENZYME databank, but to the term kinase in the Swiss-Prot databank. The SSM can match filler mappings to the appropriate function mapping via the databank attribute in SSM objects. In this manner, the terms in the ontology may be mapped to different terms appearing in the resources.
7.4 RELATED WORK
7.4.1 Information Integration in Bioinformatics
The difficulties associated with obtaining effective access to multiple biological information resources have long been recognized, and several different approaches have been proposed, making use of widely varying underlying technologies.
Probably the most widely used source integration environment for bioinformatics resources is the Sequence Retrieval Service (SRS) [16] (presented in Chapter 5). SRS is a system designed to integrate flat file databanks, which are the most common data storage form used for bioinformatics resources. SRS has its own proprietary data description and processing language. This is used to parse the flat file entries and create indices over fields and their contents. SRS has a query language for selecting entries or part of entries via Boolean combinations of indexed fields and their values. The language contains operators that can take advantage of the heavy cross-linking between different databanks. SRS is usually accessed via a Web-based interface behind which the construction of queries is hidden. The Web interface also offers supplementary analyses such as similarity and pattern scans over protein or nucleic acid sequences. SRS makes no attempt to reconcile any semantic heterogeneity between the different resources during query execution. Once results have been retrieved, the user can follow hyperlinks between entries and much use is made of this query by navigation style.
Although SRS is successful at providing navigational access between diverse resources, it provides limited facilities to support querying or programming over diverse sources. Several proposals have been made in these directions. In terms of query-oriented access, Kleisli [15] provides both a query language for ranging over data types described using a rich hierarchical data model and a collection of wrappers (known in Kleisli as drivers) for accessing biological resources. However, Kleisli has no global schema providing a model of the available data and thus can be seen as providing lower-level access to biological resources than TAMBIS. In fact, as already described, TAMBIS generates Kleisli programs as output. Another query-oriented approach is provided by the Object Protocol Model (OPM) [17], in which queries can be written over an object-oriented global model using an object query language, and tools have been developed to assist in the creation of OPM views over heterogeneous sources. The main factor that differentiates TAMBIS from OPM, from a users’ point of view, is that in TAMBIS queries are constructed over an ontology rather than over an object model. The impact of the ontology and its reasoning services on query building in TAMBIS has been discussed in Section 7.2. The ontology shields the user from the query language used, the heterogeneity of the resources, and any demand for knowledge of the resources. Such transparency may not be to all users’ tastes and more intricate queries or programs could be hand-crafted in systems such as Kleisli or OPM. Other proposals describing query-based access from object models to biological data include ISYS [18], DiscoveryLink [19] and P/FDM [20]. Kleisli, DiscoveryLink, and P/FDM are respectively presented in Chapters 6, 11, and 9.
Considerable attention has been given in bioinformatics to wrapping sources, thereby providing syntactically consistent access from programming languages to diverse resources. The bioPerl initiative1 offers a collection of Perl modules that provide access to computational techniques and data commonly found within bioinformatics resources. In the early stages of the first TAMBIS version, however, there was much interest in using the Common Object Request Broker Architecture (CORBA) to wrap bioinformatics resources [21]. CORBA allows development of object views of heterogeneous and distributed resources, regardless of their host platform, operating system, or storage paradigm. The use of CORBA within bioinformatics is promoted by the Life Sciences Research (LSR) group of the Object Management Group.2 The LSR aims to promote standard descriptions of object interfaces that enable interoperation between distributed bioinformatics resources. Among others, the European Bioinformatics Institute has provided CORBA servers for some of their databases [22]. Recently, access to SRS [16] has been provided through CORBA [23]. This service allows objects representing databank entries to be retrieved through the SRS query language. This should allow remote access to a large number of databanks and analysis programs, along with a rudimentary query facility. TAMBIS has a very different emphasis from the middleware approaches in that interactive user access is the main emphasis and in that individual sources are essentially hidden from the user in TAMBIS.
Unfortunately, the required large number of consistent, CORBA wrapped sources did not arrive to be taken advantage of by TAMBIS. The ability to download a description of a service’s interface and automatically generate a client that could act as a wrapper was desirable, but not delivered. Many providers balked at the effort needed to provide a CORBA solution to delivering services. Simple Object Protocol Servers (SOAP) servers and Web services3 offer a lighter weight solution to delivering bioinformatics services. A SOAP server for a resource is relatively cheap to set up because an object model does not have to be designed and implemented, as in CORBA. The operations available through that server can be described in the Web services description language (WSDL),4 and this description can be compiled into a client for the SOAP server. The idea is much the same as that for CORBA, but as a lighter weight solution it relies on simple message passing, not on a heavyweight object approach. These services transfer their data in extensible markup language (XML) and thus can take advantage of widely adopted XML data formats such as the biopolymer markup language [24] and the Bioinformatics Sequence Markup Language (BSML).5 XML is also seen as the data format of choice by the Interoperable Informatics Infrastructure Consortium (I3C),6 which aims to promote standards for protocols and exchange formats. The distributed annotation system (DAS)7 uses many of these ideas to manage sequence annotations distributed around the network, and delivered by SOAP servers providing an XML description of sequence annotations that allows many annotators to form an integrated, yet varied, view on the biological sequence. These technologies offer a middleware solution to the integration of bioinformatics resources. Vital though such technologies undoubtedly are, they can be seen as plumbing resources together. Choice of resources, locating those resources, knowing how to reconcile their view of the data, and the order in which to use them is still left up to the user of these technologies. TAMBIS, on the other hand, sits upon these middleware technologies and uses the ontology to offer full transparency in query management to the user.
7.4.2 Knowledge Based Information Integration
TAMBIS is one of several systems that uses a knowledge base as a central component in information integration; although, it is the first such system to be used in bioinformatics. A survey of knowledge-based information integration is given in Paton et al.’s article in Information and Software Technology [25].
In common with single interface to multiple sources (SIMS) [26], Information Manifold [27] and Observer [28], TAMBIS uses a description logic [7] to describe the concepts over which queries are to be expressed. A description logic is a modeling notation that supports reasoning over descriptions of concepts and their relationships. Two principal approaches are used in information integration systems to relate concepts in a global schema to the schemas of individual sources, namely global as view and local as view [29]. In the former, the global schema is defined as a view over the constructs in the schemas of the individual sources; in the latter the constructs in the schemas of the individual sources are defined as a view of those in the global schema. SIMS and Observer essentially use global as view techniques for processing queries, whereas Information Manifold is local as view. TAMBIS follows the global as view approach, but it generally differs from other such approaches in that very few assumptions are made of the query processing capabilities of the individual sources. In fact, as is generally true in bioinformatics, TAMBIS assumes that individual sources lack declarative query interfaces and instead provide rather limited call interfaces, supporting tasks such as iterating through the data items of a particular type or retrieving all data items with a given value for a particular attribute.
A further important feature of TAMBIS is that it supports a distinctive user interface driven from the ontology, which guides the user through the query formulation process in a way that makes it difficult to construct biologically meaningless queries. Other knowledge-based information integration systems lack such sophisticated query formulation interfaces.
7.4.3 Biological Ontologies
The number of ontologies used in bioinformatics applications is still quite small, but it is growing. However, where ontologies have been used, they span a wide range of purposes, subject areas, and representation styles [5]. The uses of bio-ontologies fall into two distinct areas: database schema definition (e.g., EcoCyc and RiboWeb) and annotation and communication (e.g., GO and OMB). The TAMBIS ontology adds a third use, ontology-based search and query formulation, to this list. The version of TAMBIS described in this chapter was the first ontology solution based on Description Logic of its type in the bioinformatics arena.
RiboWeb [30] is an ontology of ribosome structure, components, and experimental methods used to drive a Web interface that supports the analysis of ribosomal data. The ontology acts as a schema, driving the acquisition of instances that create the knowledge base. The knowledge held in the ontology also drives the analysis of new data, guiding the user as to which analysis methods are appropriate for the data in hand and indicating results that contradict current knowledge.
EcoCyc [31] uses an ontology to create an encyclopedia of E.coli metabolism, regulation, and signal transduction. As with RiboWeb, this ontology acts as a schema for the knowledge base, capturing the domain knowledge with high fidelity. Both these systems use a frame-based knowledge representation language in which a frame represents a concept and slots within frames represent attributes or roles and their fillers. Such representations can be expressive, hence the richness of the models.
The Ontology for Molecular Biology (OMB) [32] provides a framework for describing computational methods, database representations, and core molecular biological concepts. The OMB is aimed at providing a reference ontology to improve community-wide communication. Data resources would use the OMB to define their classes, relationships, and terms. The OMB uses an object-like structure with an is a kind of hierarchy and large use of other relationship types.
The Gene Ontology (GO) [33] is a structured, controlled vocabulary used to annotate gene products for their function, ultimate cellular location, and the processes in which they take part. GO is used in several genomic databases and thus adds consistency across these resources. As a consequence, querying these resources becomes more reliable. The ontology has a simple structure, relying on an is-a-kind-of hierarchy and a sparse partonomy to relate natural language phrases. The ImMunoGeneTics ontology holds terminology on the areas of immunoglobulins and their genetics. Again, this acts as a controlled vocabulary, but it has a less well-defined structure than GO and appears more like a glossary with inter-related entries.
Although all these resources can be termed ontologies, they fall into a spectrum of expressivity and formality. The frame-based systems are relatively rich, expressive, and formal, whereas the phrase-based terminologies are simpler and less expressive. The first TAMBIS ontology was the first bio-ontology to use a description logic as its representation and, as a consequence, has a more well-defined semantics than the other representations used in bio-ontologies. In contrast to the narrow range of ontology use, however, the scope and detail of the content of these ontologies varies enormously. The ImMunoGeneTics, RiboWeb, and EcoCyc ontologies are highly detailed but highly specialized to one subject area, leaving only some commonality for core areas such as gene and protein. The OMB is wide ranging and high level, whereas GO lacks any high-level conceptualization but becomes very detailed, starting its conceptualization where the OMB finishes. As has been seen, the TAMBIS ontology is broad in its conceptualization, using an upper-level ontology in which to place these concepts. The ontology is relatively shallow, but detail may be added as the user dynamically creates new concepts as compositions of pre-existing concepts and has them automatically checked and classified by the ontology’s reasoning service.
7.5 CURRENT AND FUTURE DEVELOPMENTS IN TAMBIS
The first version of TAMBIS was successful. It is possible to use an ontology describing a complex domain, such as molecular biology and bioinformatics, and use it to give the illusion of a common query interface to multiple, diverse, and heterogeneous information sources. The ontology drove a query formulation interface that allowed users to create complex queries over those multiple sources—queries that would usually need a program written by a trained bioinformatician. Usage of TAMBIS did, however, reveal some issues that needed to be addressed in further work.
Total transparency is not always desirable. The level of transparency offered by the first version of TAMBIS was appreciated by less skilled users who were happy to have decisions on which resources to use taken out of their hands. However, some users, usually those well versed in using bioinformatics resources, wished to express preferences about which resources to use, given that some sources may be more trusted than others. As the number of resources available within TAMBIS increases, such preferences will be able to be expressed. In addition, users may wish to record when and where data they retrieved arose [34]. In version one, the CPL query plan implicitly recorded some such information in the names of functions used in the query plan. It is more desirable, however, to record query provenance directly and explicitly.
The TAMBIS user survey [2] revealed that user intervention during query execution and inspection of intermediate results was desirable. Users often wish to monitor the progress of a complex, multi-source query, inspecting results to evaluate validity of the query so far and to edit data before it proceeds into subsequent parts of the query. Code for managing the execution of a query will have to be included into the code generated by the updated query processor in TAMBIS.
In the first version of TAMBIS, both the SSM and wrappers were hand-crafted. It is an aim of future versions of TAMBIS to build tools to support this process. Concepts in the ontology have to be related to methods or functions in the wrappers and information about argument and return types, and costs recorded. The ontology itself, through the ontology server, could drive such a tool and also help to check that the content of the ontology is covered within the SSM.
In the new version of TAMBIS, the TAMBIS ontology has been remodeled using the DL language DAML+OIL8 and classified using the FaCT reasoner [35], which is considerably more powerful than GRAIL used in the original TAMBIS ontology.9 This allows the biological domain to be described more precisely in the ontology and allows more precise questions to be asked by the users. In addition, the reasoning services of the DL are used extensively during query processing to support semantic query optimization based on axioms within the ontology. The query processing in version one of TAMBIS was somewhat limited, and these limitations include:
1. The ontology is represented using a relatively inexpressive DL in which certain features of the biological domain are difficult to express.
2. The mapping between concepts in the ontology and collections in the sources is quite restrictive. For example, TAMBIS did not allow multiple sources for the same kind of data (e.g., both Swiss-Prot and the Protein Information Reserve (PIR) as protein sources.
3. Although queries are optimized [13], there is no semantic query optimization making use of axioms from the ontology.
In the second version of TAMBIS, an object-oriented wrapper layer has been adopted to replace that provided by CPL and BioKleisli. Instead of a CPL query plan, a Java program is written. The use of an object-oriented wrapper layer will make TAMBIS compatible with mainstream middleware proposals such as that standardized by the Object Management Group (OMG), which in turn is associated with an important standardization activity in bioinformatics.10
7.5.1 Summary
This chapter has provided an overview of the first TAMBIS system for querying distributed bioinformatics sources. The key contributions of TAMBIS are:
1. It is the first ontology-based information integration system to be used in bioinformatics. Although ontologies are becoming important in bioinformatics for annotating databases [33] and for managing complex information resources [30], TAMBIS is the first project to use ontologies to support the important task of integrating bioinformatics resources.
2. TAMBIS is centered on the first description logic-based ontology in bioinformatics. Other ontologies in bioinformatics have made use of frame-based representations or structured terminologies, but they are not amenable to subsumption reasoning as in TAMBIS.
3. The user interface in TAMBIS is driven directly from the ontology, and as such, it both guides the user in constructing well formed requests and detects when biologically nonsensical questions have been asked. Other knowledge-based information integration systems have paid less attention to user interaction issues.
4. The TAMBIS query processor has been integrated with existing wrapping software, allowing re-use of established middleware techniques and existing wrappers. The query processor makes minimal assumptions on the query interfaces made available by sources, reflecting the limited public query interfaces generally available in bioinformatics.
TAMBIS seeks to provide correct answers to precisely formed queries. Queries can be expressed precisely, at a level of detail corresponding to that of the underlying resources, by using the ontology to constrain what it is valid to ask. Answers should be correct because the sources and services model makes explicit how queries expressed over the ontology can be answered using the available sources. However, such quality of service is achieved at some cost; the development of ontologies that describe a domain is a skilled and time-consuming process (the 1800-concept TAMBIS ontology took 2 person-years to write), and incorporating a wrapped source into the SSM is itself a manual and time-consuming task. However, these two tasks involve (1) describing what it is valid to ask of a collection of bioinformatics sources and (2) describing how to obtain answers from a collection of sources. Although the developers and maintainers of a TAMBIS installation must undertake these tasks, the users of the TAMBIS system need not, and thus they can benefit from the knowledge encoded in the ontology and in the SSM.
ACKNOWLEDGMENTS
This work is funded by AstraZeneca pharmaceuticals, the BBSRC/EPSRC Bioinformatics Initiative (grant number BIF/05344), and the EPSRC Distributed Information Management Initiative (grant number GR/M76607), whose support we are pleased to acknowledge. We are also grateful to Alex Jacoby and Martin Peim for their contributions to the implementation of the TAMBIS system.
REFERENCES
[1] Markowitz, V.M., Ritter, O. Characterizing Heterogeneous Molecular Biology Database Systems. Journal of Computational Biology. 1995;2(no. 4):547–556.
[2] Stevens, R.D., Goble, C.A., Baker, P., et al. A Classification of Tasks in Bioinformatics. Bioinformatics. 2001;17(no. 2):180–188.
[3] Goble, C.A., Stevens, R., Ng, G., et al. Transparent Access to Multiple Bioinformatics Information Sources. IBM Systems Journal. 2001;40(no. 2):532–552.
[4] Buneman, P., Davidson, S.B., Hart, K., et al. A Data Transformation System for Biological Data Sources. In: Proceedings of the 21st International Conference on Very Large Data Bases (VLDB). San Francisco: Morgan Kaufmann; 1995:158–169.
[5] November Stevens, R., Goble, C.A., Bechhofer, S. Ontology-Based Knowledge Representation for Bioinformatics. Briefings in Bioinformatics. 2000;1(no. 4):398–416.
[6] Baker, P.G., Goble, C.A., Bechhofer, S., et al. An Ontology for Bioinformatics Applications. Bioinformatics. 1999;15(no. 6):510–520.
[7] Borgida., A. Description Logics in Data Management. IEEE Transactions on Knowledge and Data Engineering. 1995;7(no. 5):785–798.
[8] Ringland, G.A., Duce, D.A. Approaches to Knowledge Representation: An Introduction. New York: John Wiley; 1988.
[9] Baader F., McGuinness D., Nardi D., et al, eds. The Description Logic Handbook: Theory, Implementation and Applications. Cambridge, UK: Cambridge University Press, 2003.
[10] Rector, A.L., Bechhofer, S.K., Goble, C.A., et al. The GRAIL Concept Modelling Language for Medical Terminology. Artificial Intelligence in Medicine. 1997;9(no. 2):139–171.
[11] Donini, F.M., Lenzerini, M., Nardi, D., et al. Reasoning in Description Logics. In: Foundations of Knowledge Representation. Stanford, CA: Center for the Study of Language and Information (CSLI) Publications; 1996:191–236.
[12] Bechhofer, S., Stevens, R., Ng, G., et al. Guiding the User: An Ontology Driven Interface. In: Paton N.W., Griffiths T., eds. Proceedings of User Interfaces to Data Intensive Systems (UIDIS99). New York: IREE Press; 1999:158–161.
[13] Paton, N.W., Stevens, R., Baker, P., et al. Query Processing in the TAMBIS Bioinformatics Source Integration System. In: Proceedings of the 11th International Conference on Scientific and Statistical Database Management (SSDBM). New York: IEEE Press; 1999:138–147.
[14] Swami, A.N. Optimization of Large Join Queries. In: Proceedings of the 1989 ACM SIGMOD International Conference on Managing Error. New York: ACM Press; 1989:367–376.
[15] November Davidson, S.B., Overton, C., Tannen, V., et al. BioKleisli: A Digital Library for Biomedical Researchers. Journal of Digital Libraries. 1997;1(no. 1):36–53.
[16] Ezold, T., Ulyanov, A., Nardi, D., et al. SRS: Information Retrieval System for Molecular Biology Data Banks. Methods in Enzymology. 1996;266:114–128.
[17] Chen, I-M.A., Kosky, A.S., Markowitz, V.M., et al. Constructing and Maintaining Scientific Database Views in the Framework of the Object Protocol Model. In: Proceedings of the 9th International Conference on SSDBM. New York: IEEE Press; 1997:237–248.
[18] Siepel, A.C., Tolopko, A.N., Farmer, A.D., et al. An Integration Platform for Heterogeneous Bioinformatics Software components. IBM Systems Journal. 2001;40(no. 2):570–591.
[19] Haas, L.M., Kodali, P., Rice, J.E., et al, Integrating Life Sciences Data with a Little Garlic, Proceedings of the International Symposium on Bio-Informatics and Biomedical Engineering (BIBE). New York: IEEE Press; 2000;5–12.
[20] Kemp, G.J.L., Angelopoulog, N., Gray, P.M.D. A Schema-Based Approach to Building a Bioinformatics Database Federation. In: Proceedings of the International Symposium on Bio-Informatics and Biomedical Engineering (BIBE). New York: IEEE Press; 2000:13–20.
[21] Stevens, R., Miller, C. Wrapping and Interoperating Bioinformatics Resources Using CORBA. Briefings in Bioinformatics. 2000;1(no. 1):9–21.
[22] Rodriguez-Tomé, P., Helgesen, C., Lijnzaad, P., et al. A CORBA Server for the Radiation Hybrid DataBase. In: Proceedings of the Fifth International Conference on Intelligent systems for Molecular Biology. Menlo Park, CA: AAAI Press; 1997:250–253.
[23] Coupaye, T. Wrapping SRS With CORBA: From Textual Data to Distributed Objects. Bioinformatics. 1999;15(no. 4):333–338.
[24] Fenyo, D. The Biopolymer Markup Language. Bioinformatics. 1999;15(no. 4):339–340.
[25] Paton, N.W., Goble, C.A., Bechhofer, S. Knowledge Based Information Integration Systems. Information and Software Technology. 2000;42(no. 5):299–312.
[26] Arens, Y., Knoblock, C.A., Shen, W-M. Query Reformulation for Dynamic Information Integration. Journal of Intelligent Information Systems. 1996;6(no.2–3):99–130.
[27] Levy, A.Y., Srivastava, D., Kirk, T. Data Model and Query Evaluation in Global Information Systems. Journal of Intelligent Information Systems. 1995;5(no. 2):121–143.
[28] Mena, E., Illarramendi, A., Kashyap, V., et al. Observer: An Approach for Query Processing in Global Information Systems Based on Interoperation Across Pre-Existing Ontologies. Distributed and Parallel Databases. 2000;8(no. 2):223–271.
[29] Ullman, J.D. Information Integration Using Logical Views. In: Proceedings of ICDT ’97: 6th International Conference on Database Theory. Heidelberg, Germany: Springer-Verlag; 1997:19–40.
[30] Altman, R., Bada, M., Chai, X.J., et al. RiboWeb: An Ontology-Based System for Collaborative Molecular Biology. IEEE Intelligent Systems. 1999;14(no. 5):68–76.
[31] Karp, P., Riley, M., Paley, S., et al. EcoCyc: Electronic Encyclopedia of E. coli Genes and Metabolism. Nucleic Acids Research. 1999;27(no. 1):55–58.
[32] Schulze-Kremer, S. Ontologies for Molecular Biology. In: Proceedings of the Third Pacific Symposium on Biocomputing. Singapore: World Scientific; 1998:693–704.
[33] Ashburner, M., Ball, C.A., Blake, J.A., et al. Gene Ontology: Tool for the Unification of Biology. Nature Genetics. 2000;25(no. 1):25–29.
[34] Buneman, P., Khanna, S., Tan, W-C., Data Provenance: Some Basic Issues, TSTTCS 2000: 29th Conference on Foundations of Software Technology and Theoretical Computer Science, New Dehli, India. Lecture Notes in Computer Science. 1974. Heidelberg, Germany: Springer-Verlag; 2000:87–93.
[35] Horrocks, I. Using an Expressive Description Logic: Fact or Fiction. In: Cohn A.G., Schubert L.K., Shapiro S.C., eds. Principles of Knowledge Representation and Reasoning: Proceedings of the Sixth International Conference (KR ’98). San Francisco: Morgan Kaufmann; 1998:636–647.
1Information about the bioPerl initiative is available at http://bioperl.org.
2Go to http://www.omg.org/lsr for information on the Life Sciences Research effort of the Object Management Group.
3The Simple Object Protocol Servers (SOAP) and Web service protocols are available at http://www.w3c.org/soap.
4For more information about the WSDL, refer to http://www.w3.org/TR/wsdl.
5The BSML is available at http://www.bsml.org.
6Refer to http://www.i3c.org for information about I3C.
7Go to http://www.biodas.org for information on biological DAS.
8Information on DAML+OIL can be found at http://www.daml.org.
9Versions of this ontology represented in DAML+OIL may be found at http://img.cs.man.ac.uk/stevens/tambis-oil.html.
10Information about the effort of standardization of bioinformatics by the OMG is available at http://www.omg.org/homepages/lsr/.










