DiscoveryLink
DiscoveryLink enables the integration of diverse data from diverse sources into a single, virtual database, with the goal of making it easier for scientists to find the information they need to prevent and cure diseases. To progress in this quest, scientists need to answer questions that relate data about genomics, proteomics, chemical compounds, and assay results, which are found in relational databases, flat files, extensible markup language (XML), Web sites, document management systems, applications, and special-purpose systems. They need to search through large volumes of data and correlate information in complex ways.
In bioinformatics research in the post-genomic era, the sheer volume of data and number of techniques available for use in the identification and characterization of regions of functional interest in the genomic sequence is increasing too quickly to be managed by traditional methods. Investigators must deal with the enormous influx of genomic sequence data from human and other organisms. The results of analysis applications such as the Basic Local Alignment Search Tool (BLAST) [1], PROSITE [2], and GeneWise [3] must be integrated with a large variety of sequence annotations found in data sources such as GenBank [4], Swiss-Prot [5], and PubMed [6]. Public and private repositories of experimental results, such as the Jackson Laboratory’s Gene Expression Database (GXD) [7], must also be integrated. Deriving the greatest advantage from this data requires full, query-based access to the most up-to-date information available, irrespective of where it is stored or its format, with the flexibility to customize queries easily to meet the needs of a variety of individual investigators and protein families.
In an industrial setting, mergers and acquisitions increase the need for data integration in the life science industry in general and the pharmaceutical industry in particular. Even without mergers, in a typical pharmaceutical company, the research groups are geographically dispersed and divided into groups based on therapeutic areas. Scientists in each of these therapeutic areas might be involved in various stages of the drug discovery process such as target identification, target validation, lead identification, lead validation, and lead optimization. During each of these stages, they need to access diverse data sources, some specific to the therapeutic area of interest and the particular stage of the process and others that are of value to many therapeutic areas and at many stages of the process. Providing the data integration infrastructure to support this research environment (geographically dispersed research groups accessing different sets of diverse data sources depending on their area of research and the stage of the drug discovery process) is a daunting task for any information technology (IT) group.
As pharmaceutical companies try to shorten the drug-discovery cycle, they must identify new drug candidates more quickly by increasing the efficiency of the research processes and eliminating the false positives earlier in the discovery process. Providing scientists with easy access to the relevant information is essential. Researchers working in the gene expression domain may gain valuable insights if they have access to data from comparative genomics, biological pathways, or cheminformatics. This is also true for a scientist working in the lead identification or optimization areas.
This case can be illustrated by an example. A research group in a pharmaceutical company working in a particular therapeutic area might be interested in looking at all the compounds active in biological assays that have been generated and tested for a given receptor. In addition, the researchers might be interested in looking at similar compounds and their activities against similar receptors. This will help them understand the specificity and selectivity of the compounds identified. The knowledge that a particular set of compounds was considered for a different therapeutic area by another team could help them develop new leads or eliminate compounds that are not specific in their activities. To answer these queries, the research group must correlate information from multiple databases, some relational (e.g., the assay data may be stored relationally), some not (the chemical structure data might be stored by a special-purpose system), and use specialized functions of the data sources (e.g., similarity searches involving compound structures or DNA sequences).
There are many different approaches to integrating diverse data sources. Often, integration is provided by applications that can talk to one of several data sources, depending on the user’s request. In these systems, access to the data sources is typically “hard-wired.” Replacing one data source with another means rewriting a portion of the application. In addition, data from different sources cannot be compared in response to a single request unless the comparison is likewise wired into the application. Moving all relevant data to a warehouse allows greater flexibility in retrieving and comparing data, but at the cost of re-implementing or losing the specialized functions of the original source, as well as the cost of maintenance. A third approach is to create a homogeneous object layer to encapsulate diverse sources. This encapsulation makes applications easier to write and more extensible, but it does not solve the problem of comparing data from multiple sources.
To return to the example, today this problem would be addressed by writing an application that accesses chemical structure databases (with specific functionality such as similarity or substructure searches), assay databases (maybe in relational format), and sequence databases (flat file, relational, or XML format). Answering the previous question requires multiple queries against these data sources:
1. “Show me all the active compounds for each of the assays for a particular receptor.”
2. “Show me all the compounds that are similar to the top five compounds from the previous query” (may require multiple requests, one per compound, depending on the sophistication of the data store and application).
3. “Do a BLAST [1] run to find similar receptors.”
4. “Show me the results of the compounds from Query 2 from all the assays against the receptors of Query 3” (may require multiple requests, one per compound or one per compound-receptor pair, depending on the sophistication of the data store and application).
5. “Sort the result set by order of the specificity or selectivity information” (if multiple queries were needed in Step 4).
Depending on the activities of the set of compounds, various scenarios emerge that tell the researchers how best to continue their research. However, the application is hard to write and may need to be extended if additional sources are needed (e.g., if a new source of compound or assay information is acquired).
A virtual database, on the other hand, offers users the ability to combine data from multiple sources in a single query without creating a physical warehouse. DiscoveryLink [8] uses federated database technology to provide integrated access to data sources used in the life sciences industry. The federated middleware wraps the actual data sources, providing an extensible framework and encapsulating the details of the sources and how they are accessed. In this way, DiscoveryLink provides users with a virtual database to which they can pose arbitrarily complex queries in the high-level, non-procedural query language SQL. DiscoveryLink focuses on efficiently answering these queries, even though the necessary data may be scattered across several different sources, and those sources may not themselves possess all the functionality needed to answer such a query. In other words, DiscoveryLink is able to optimize queries and compensate for SQL functions that may be lacking in a data source. Additionally, queries can exploit the specialized functions of a data source, so no functionality is lost in accessing the source through DiscoveryLink.
Using DiscoveryLink in the example, a single query could retrieve the structures of compounds that are active in multiple assays against different receptors. Views could be defined to create a canonical representation of the data. Furthermore, the query would be optimized for efficient execution. DiscoveryLink’s goal is to give the end user the perspective of a single data source, saving effort and frustration. In a real scenario, before researchers propose the synthesis and testing of an interesting compound they have found, they would like to know the toxicity profile of the compound and related compounds and also the pathways in which the compound or related compounds might be involved. This would require gathering information from a (proprietary) toxicity database, as well as one with information on metabolic pathways, such as the Kyoto Encyclopedia of Genes and Genomes (KEGG), and using the structures and names of the compounds to look up the data—another series of potentially tricky queries without an engine such as DiscoveryLink.
This chapter presents an overview of DiscoveryLink and shows how it can be used to integrate life sciences data from heterogeneous data sources. The next section provides an overview of the DiscoveryLink approach, discussing the data representation, query capability, architecture, and the integration of data sources, as well as providing a brief comparison to other systems for data integration. Section 11.2 focuses on query processing and optimization. Section 11.3 addresses performance, scalability, and ease of use. The final section concludes with some thoughts on the current status and success of the system, as well as some directions for future enhancements.
11.1 APPROACH
DiscoveryLink is based on federated database technology, which offers powerful facilities for combining information from multiple data sources. Built on technology from an earlier product, DB2 DataJoiner [9], and enhanced with additional features for extensibility and performance from the Garlic research project [10, 11], DiscoveryLink’s federated database capabilities provide a single, virtual database to users. DB2 DataJoiner first introduced the concept of a virtual database, which is created by federating together multiple heterogeneous, relational data sources. Users of DB2 DataJoiner could pose arbitrary queries over data stored anywhere in the federated system without worrying about the data’s location, the SQL dialect of the actual data store(s), or the capabilities of those stores. Instead, users had the full capabilities of DB2 against any data in the federation. The Garlic project demonstrated the feasibility of extending this idea to build a federated database system that effectively exploits the query capabilities of diverse, often non-relational data sources. In both of these systems, as in DiscoveryLink, a middleware query processor develops optimized execution plans and compensates for any functionality the data sources may lack.
There are many advantages of a federated database approach to integrating life science data. In particular, this approach is characterized by transparency (the degree to which it hides all details of data location and management), heterogeneity (the extent to which it tolerates data source diversity), a high degree of function providing the benefits of both SQL and the underlying data source capabilities, autonomy for the underlying federated sources, easy extensibility, openness, and optimized performance. All other approaches fall short in one or another of these categories. These other approaches are numerous, including domain-specific solutions, language-based frameworks, dictionary-based solutions, frameworks based on an object model, and data warehousing approaches.
For example, companies like Informax provide data retrieval and data integration for biological databases. Their system, and many like it, benefits from being created specifically for bioinformatics data, but as a result, it cannot readily exploit advances in query processing (e.g., in the relational database industry). Kleisli’s [12] Collection Programming Language (CPL) presented in Chapter 6 allows the expression of complicated transformations across heterogeneous data sources, but it provides no global schema, making query formulation and optimization difficult. The Sequence Retrieval System (SRS) [13, 14] presented in Chapter 5 provides fast access to a vast number of text files, and LION provides a rich biology workbench of integrated tools built on SRS. SRS has its own proprietary query language, which offers excellent support for navigational access but less power for cross-source querying than SQL. In fact, LION’s DiscoveryCenter uses DiscoveryLink to extend its database integration capabilities. Biomax provides similar functionality in its Biological Databanks Retrieval System (BioRS) tool, with cleanly structured interfaces based on the Common Object Request Broker Architecture (CORBA) for scalability on both multi-processors and workstations alike. BioRS also offers a curated and annotated database of the human genome and a number of powerful analysis tools. Again, while the domain-specific tooling makes this a great package for biologists, the language used for queries is more limited than SQL. Accelrys provides a relational data management and analysis package, SeqStore, and a rich set of bioinformatics tools for sequence analysis—the Genetics Computer Group (GCG) Wisconsin Package. SeqStore includes a relational data warehouse for sequence data, coupled with tools to receive automated updates, to analyze sequences with the wide range of analyses available in the GCG Wisconsin Package, and to create automated sequence analysis pipelines. The warehousing approach requires that data be moved (or copied), interfering with source autonomy and limiting the extensibility of the system—or at least making it harder to extend. The object frameworks, such as that provided by Tripos, provide only limited transparency. Similar arguments apply to most other bioinformatics integration engines.
The two biology-focused integration engines that come closest to DiscoveryLink’s vision are Gene Logic’s Object Protocol Model (OPM) [15] and the Transparent Access to Multiple Biological Information Systems (TAMBIS) [16], presented in Chapter 7. OPM provides a virtual, object-oriented database, with queries in the proprietary OPM-MQL query language over diverse query sources. OPM’s query optimization is rule-based and somewhat limited, because of the difficulties of optimizing over its more complex data model. While an object-oriented model is a natural choice for modeling life sciences data, and OPM’s class methods have been demonstrated to add significant scientific value [17], DiscoveryLink follows an industry standard (relational), believing that the virtues of openness and the benefits of riding on technology that is constantly evolving and growing in power (due to the large number of users and uses) outweighed the annoyances of modeling data as relations. In fact, the database industry is now rapidly adding support for XML and XQuery to its once purely relational products; DiscoveryLink will exploit these capabilities as they become available, alleviating any modeling issues substantially. For example, the DiscoveryLink engine already supports SQL/XML functions that allow it to return XML documents instead of tuples.
TAMBIS is unique in its use of an ontology to guide query formulation, query processing, and data integration. It also offers users a virtual database and deals with a great deal of heterogeneity. Originally based on CPL wrappers for accessing data sources, TAMBIS now uses a more general Java wrapper mechanism. TAMBIS focuses on supporting direct user interactions, unlike DiscoveryLink, which is meant to be a general infrastructure against which many different query tools can be used. Again, DiscoveryLink benefits from its open, industry-standard interfaces for both queries and wrappers. However, the use of an ontology to generate and refine queries is a powerful mechanism, and the marriage of such techniques to DiscoveryLink middleware could be explored to provide a more biology-centric experience for users.
Because DiscoveryLink is a general platform for data integration, it also can be compared to other database integration offerings. Most of the major database vendors offer some sort of cross-database query product, often called a gateway. For example, Oracle offers both dblinks (for cross-Oracle queries) and Oracle Transparent Gateway (for more heterogeneous data sources). DiscoveryLink differs from these and other products in three fundamental ways: (1) It offers an open application programming interface (API) for wrapper construction; (2) it allows the use of data source functions in queries that span multiple data sources; and (3) it has the most powerful optimization capabilities available (it is the only system that takes query-specific input from wrappers during query planning). Few systems offer the same degree of transparency and the same query processing power against heterogeneous sources.
11.1.1 Architecture
The overall architecture of DiscoveryLink, shown in Figure 11.1, is common to many heterogeneous database systems, including the Stanford-IBM Manager of Multiple Information Sources (TSIMMIS) [18], Distributed Information Search Component (DISCO) [19], Pegasus [20], Distributed Interoperable Object Model (DIOM) [21], Heterogeneous Reasoning and Mediator System (HERMES) [22], and Garlic [10, 11]. Applications connect to the DiscoveryLink server using any of a variety of standard database client interfaces, such as Call Level Interface (CLI) [23], Object Database Connectivity (ODBC), or Java Database Connectivity (JDBC), and submit queries to DiscoveryLink in standard SQL (specifically SQL3 [24]). The information required to answer the query comes from the local database and/or from one or more data sources, which have been identified to DiscoveryLink through a process called registration. The data from the sources is modeled as relational tables in DiscoveryLink. The user sees a single, virtual relational database, with the original locations and formats of the sources hidden. The full power of SQL is supported against all the data in this virtual database, regardless of where the data is actually stored and whether the data source actually supports the SQL operations.
When an application submits a query to the DiscoveryLink server, the server identifies the relevant data sources and develops a query execution plan for obtaining the requested data. The plan typically breaks the original query into fragments that represent work to be delegated to individual data sources and additional processing to be performed by the DiscoveryLink server to filter, aggregate, or merge the data. The ability of the DiscoveryLink server to further process data received from sources allows applications to take advantage of the full power of the SQL language, even if some of the information they request comes from data sources with little or no native query processing capability, such as simple text files. The local data store allows query results to be stored for further processing and refinement, if desired, and also provides temporary storage for partial results during query processing.
The DiscoveryLink server communicates with a data source by means of a wrapper [11], a software module tailored to a particular family of data sources. The wrapper for a data source is responsible for four tasks:
![]() Mapping the information stored by the data source into DiscoveryLink’s relational data model
Mapping the information stored by the data source into DiscoveryLink’s relational data model
![]() Informing DiscoveryLink about the data source’s query processing capabilities by analyzing plan fragments during query optimization
Informing DiscoveryLink about the data source’s query processing capabilities by analyzing plan fragments during query optimization
![]() Mapping the query fragments submitted to the wrapper into requests that can be processed using the native query language or programming interface of the data source
Mapping the query fragments submitted to the wrapper into requests that can be processed using the native query language or programming interface of the data source
The interface between the DiscoveryLink server and the wrapper supports the International Standards Organization/Structured Query Language/Management of External Data (ISO SQL/MED) standard [25].
Wrappers are the key to extensibility in DiscoveryLink, so one of the primary goals for the wrapper architecture was to allow wrappers for the widest possible variety of data sources to be produced with a minimum of effort. Past experience has shown that this is feasible. To make the range of data sources that can be integrated using DiscoveryLink as broad as possible, a data (or application) source only needs to have some form of programmatic interface that can respond to queries and, at a minimum, return unfiltered data that can be modeled (by the wrapper) as rows of (one or more) tables. The author of a wrapper need not implement a standard query interface that may be too high-level or low-level for the underlying data source. Instead, a wrapper provides information about a data source’s query processing capabilities and specialized search facilities to the DiscoveryLink server, which dynamically determines how much of a given query the data source is capable of handling. This approach allows wrappers for simple data sources to be built quickly, while retaining the ability to exploit the unique query processing capabilities of non-traditional data sources such as search engines for chemical structures or images. For DiscoveryLink, this design was validated by wrapping a diverse set of data sources including flat files, relational databases, Web sites, a specialized search engine for text, and the BLAST search engine.
To make wrapper authoring as simple as possible, only a small set of key services from a wrapper is required, and the approach ensures that a wrapper can be written with very little knowledge of DiscoveryLink’s internal structure. As a result, the cost of writing a basic wrapper is small. In past experience, a wrapper that just makes the data at a new source available to DiscoveryLink, without attempting to exploit much of the data source’s native query processing capability, can be prototyped in a matter of days by someone familiar with the data source interfaces and the wrapper concepts. Because the DiscoveryLink server can compensate for missing functionality at the data sources, even such a simple wrapper allows applications to apply the full power of SQL to retrieve the new data and integrate it with information from other sources, albeit with perhaps less-than-optimal performance. Once a basic wrapper is written, it can be incrementally improved to exploit more of the data source’s query processing capability, leading to better performance and increased functionality as specialized search algorithms or other novel query processing facilities of the data source are exposed.
A DiscoveryLink wrapper is a C++ program, packaged as a shared library that can be loaded dynamically by the DiscoveryLink server when needed. Often a single wrapper is capable of accessing several data sources, as long as they share a common or similar API. For one thing, wrappers do not need to encode information on the schema of data in the source. For example, the Oracle wrapper provided with DiscoveryLink can be used to access any number of Oracle databases, each having a different schema. In fact, the same wrapper supports several Oracle release levels as well. This has a side benefit, namely that schemas can evolve without requiring any change in the wrapper as long as the source’s API remains unchanged. In addition, wrappers can get connection information for individual servers from SQL Data Definition Language (DDL) statements, even if the schemas are identical. On the other hand, there is a tradeoff between flexibility and ease of configuration (the more flexible the wrapper, the more it needs to be told during registration). For that reason, it is sometimes more practical to encode (parts of) the schema in the wrapper. For example, the BLAST wrapper defines many fixed columns, but allows the user to specify others that are appropriate for their instantiation of BLAST.
This architecture has many benefits, as described previously. However, there are some controversial aspects. First and foremost, much biology data is semi-structured, and the current implementation forces data to be modeled relationally. While this does complicate wrapper writing somewhat, there are several examples of wrappers today that deal with nested and semi-structured data, including an XML wrapper. These wrappers expose their data as multiple relations, which can be joined to get back the full structure (note that the data is still stored in its nested form, and the joins are often translated into simple retrievals as a result). Future direction is to support XML and XQuery natively in DiscoveryLink’s engine and to allow wrapper writers their choice of a relational or an XML model. That will make the modeling issues less painful.
A second issue is the use of C++, a general purpose and somewhat arcane programming language, for writing wrappers, as opposed to a simpler scripting language or a specialized wrapper construction mechanism. A general-purpose language was chosen for several reasons. First, DiscoveryLink is meant to handle large-scale queries over many data sources and large volumes of data. C++ is an efficient language, suitable for such applications. Second, DiscoveryLink wrappers are required to do more than ordinary connectors or adaptors, and the general-purpose programming language allows the wrapper writer complete flexibility in accomplishing the wrapper tasks. A toolkit and tools for wrapper development can ease the pain of programming by providing template functions, automatic generation of parts of the code, error checking, and so on. Last but not least, the DiscoveryLink engine happens to be written in C++, so this was by far the easiest to interface with the engine initially. A Java version of the toolkit is currently produced, as well as a set of generic wrappers for particular styles of data source access (e.g., a Web services wrapper, ODBC and JDBC wrappers, maybe even a Perl script wrapper). These facilities should increase the ease of adding new wrappers.
Related to the ease of wrapper writing is the ease of changing wrappers when (if) the interface to the data source changes. For most data sources, such changes are uncommon (note that this is not in reference to schema changes but to changes in the API or language used by the data source). When changes do occur, they are often additions to the existing interface, and the wrapper can continue to function as-is, only needing modification if exploiting the new feature(s) is desired. Most commercial data sources, for example, try to maintain upward compatibility in interface between one release and the next. But for some classes of sources (especially Web data sources), change is much more common. To deal with these sources, it is particularly desirable to have some non-programmatic or scripted way of creating wrappers. Our explorations into generic wrappers that can be easily tailored will address this concern.
11.1.2 Registration
The process of using a wrapper to access a data source begins with registration, the means by which a wrapper is defined to DiscoveryLink and configured to provide access to selected collections of data managed by a particular data source. Registration consists of several steps, each taking the form of a DDL statement. Each registration statement stores configuration meta-data in system catalogs maintained by the DiscoveryLink server.
The first step in registration is to define the wrapper itself and identify the shared library that must be loaded before the wrapper can be used. The CREATE WRAPPER statement serves this purpose. BLAST [1] is a search engine for finding nucleotide or peptide sequences similar to a given pattern sequence. A wrapper for BLAST might be created as follows:
![]()
Note that a particular data source has not yet been identified, only the software required to access any data source of this kind. The next step of the registration process is to define specific data sources using the CREATE SERVER statement. If several sources of the same type are to be used, only one CREATE WRAPPER statement is needed, but a separate CREATE SERVER would be needed for each source. For a particular BLAST service, the statement might be as follows:

This statement registers a data source that will be known to DiscoveryLink as TBlastNServ and indicates that it is to be accessed using the previously registered wrapper, BlastWrapper. It further identifies that this BLAST server is doing a tBLASTn search (i.e., comparing an amino acid sequence, the input, to a database of nucleotide sequences) and that it is using version 2.1.2 of the BLAST software. The additional information specified in the OPTIONS clause is a set of pairs (option name, option value) that are stored in the DiscoveryLink catalogs but are meaningful only to the relevant wrapper. In this case, they indicate to the wrapper that the TBlastNServ data source can be contacted via a particular Internet Protocol (IP) address and port number. In general, the set of valid option names and option values will vary from wrapper to wrapper because different data sources require different configuration information. Options can be specified on each of the registration DDL statements and provide a simple but powerful form of extensible meta-data. Because a wrapper understands the options it defines, only that wrapper can validate that the option names and values specified on a registration statement are meaningful and mutually compatible. As a result, wrappers participate in each step of the registration process and may reject, alter, or augment the option information provided in the registration DDL statement.
The third registration step is to identify, for each data source, particular collections of data that will be exposed to DiscoveryLink applications as tables. This is done using the CREATE NICKNAME statement. Collectively, these statements define the schema of each data source and form the basis of the integrated schema seen by applications.
For example, suppose there are three data sources. One is a relational database system providing data on protein targets. The second is a Web site storing information about technical publications. The third is a BLAST server that has the ability to compare an input sequence to a file of stored sequences as described previously. For this example, three sets of CREATE NICKNAME statements are needed, one set for each of the three data sources. Figure 11.2 shows representative CREATE NICKNAME statements that define partial schemas for each source.
The protein sequence source exports two relations. The first is Proteins, with columns representing the unique identifier for a protein, the common (print) name, the amino acid sequence associated with the protein, the function of the protein, and a list of diseases with which the protein has been associated. In real life, a Database Administrator (DBA) would likely declare a fuller set of columns, representing more of the information contained in the source; the schema is simplified in the interest of space only. Also, because the data source—a relational Database Management System (DBMS)—has a self-describing schema, the DBA would not actually need to put the column information in the CREATE NICKNAME statement. That information could be read from the data source catalogs automatically. The second relation exported from this source is a mapping table that maps proteins to publications that reference them. The FOR clause identifies, via a three-part name, the server, schema, and remote table referenced by the nickname. This syntax may be used with relational data sources.
Similarly, the DBA makes visible a single table, Pubs, from the publication source, for which only four columns are shown: the publication identifier, the title of the article, the date the article was published, and a list of keywords for the publication. Note that the nickname definitions give the types of attributes in terms of standard SQL data types. This represents a commitment on the part of the wrapper to translate types used by the data source to these types as necessary.
Finally, the BLAST search engine is modeled as a virtual table, indexed on the input sequence and with columns representing both input parameters and the results of the BLAST search. Again, only a subset of the schema is shown. Here are shown the input column, query_seq, and output columns for the accession number, definition, and hsp_info (the information string computed for a given high-scoring segment pair containing information about the number of nucleotides or amino acids that matched between the query and the hit sequences). Note the use of options clauses on both the CREATE NICKNAME statement and on the definition of individual columns. These give the DBA the ability to specify information needed by the wrapper. For the BLAST wrapper, the options on the individual columns tell the wrapper how to parse the BLAST defline into these columns. In this case, the defline is assumed to contain the accession number, followed by the definition, delimited by white space. (Columns whose values do not come from the defline have no parsing options specified.) The option on the overall CREATE NICKNAME tells the wrapper which data source to blast against (in this case GenBank’s gbest). Actually, the BLAST wrapper supports so many different input and output columns that part of the schema is hard-wired so a DBA does not have to re-type all the columns in the CREATE NICKNAME statement. Further details on this wrapper can be found in the IBM DB2 Life Sciences Data Connect Planning, Installation and Configuration Guide [26].
Specialized search or data manipulation capabilities of a data source also can be modeled as user-defined functions, and identifying these functions by means of CREATE FUNCTION MAPPING statements is the fourth step in registration. Thus, the definition of the publications data source in Figure 11.2 also includes a CREATE FUNCTION MAPPING statement, registering that source’s function contains (A, B, C). This function returns ‘Y’ if the publication identified by A contains the string C in column B, for example, contains (‘ML546’, ‘keywords’, ‘ovarian cyst’). The mapping identifies this function to the query processor and declares its signature and return type in terms of standard SQL data types. As with nicknames, the wrapper must convert values of these types to and from the corresponding types used by the data source. This function models the underlying data source’s Boolean search capability.
Finally, user mappings are defined. A user mapping tells DiscoveryLink how to connect a particular local user to a data source. For example, if a DiscoveryLink user identified by LAURA connects to the protein database as ITNerd, using the password DLRocks, the following DDL statement might be issued:
![]()
With these five steps, registration is complete. The new data source is ready to use. Queries can combine data from all the registered sources and use the specialized capabilities of these sources; in the example, two techniques for modeling these special capabilities were shown: using a virtual table, as done for the BLAST source, and using a function mapping, as done for the contains function of the publications source. Note that additional sources can be added at any time without affecting the ongoing operations of the federated system. The system need not be quiesced, and existing applications and queries need not be altered. However, new queries that combine information from the preexisting sources and the new source can now be asked.
If data source schemas or functions change, they must be re-registered. DiscoveryLink currently has no mechanism to detect changes in the sources, though an application that periodically compares the DiscoveryLink and source schemas could be written.
11.2 QUERY PROCESSING OVERVIEW
Once registration is completed, the newly defined nicknames and functions can be used in queries. When an application issues a query, the DiscoveryLink server uses the meta-data in the catalogs to determine which data sources hold the requested information. Then it optimizes the query, looking for an efficient execution plan. It explores the space of possible query plans, using dynamic programming to enumerate plans for joins. The optimizer first generates plans for single table accesses, then for two-way joins, and so on. With each round of planning, the optimizer considers various join orders and join methods, and if all the tables are located at a common data source, it tries to generate plans for performing the join either at the data source or at the federated server.
Once the optimizer has chosen a plan for a query, query fragments are distributed to the data sources for execution. Each wrapper maps the query fragment it receives into a sequence of operations that make use of its data source’s native programming interface and/or query language. Once the plan has been translated, it can be executed immediately or saved for later execution. The DiscoveryLink server’s execution engine is pipelined and employs a fixed set of functions (open, fetch, close) that each wrapper must implement to control the execution of a query fragment. When accepting parameters from the server or returning results, the wrapper is responsible for converting values from the data source type system to DiscoveryLink’s SQL-based type system.
DiscoveryLink includes a full database engine that can execute arbitrary (DB2) SQL queries. Features useful for life sciences applications include support for long data types (e.g., Binary Large Object [BLOB], Character Large Object [CLOB]) and user-defined functions. Applications also benefit from the ability to update information at relational data sources via SQL statements submitted to DiscoveryLink (and in the future, full transaction management for data sources that comply with the X/Open XA-interface specification), the ability to invoke stored procedures that reference nicknames, and the ability to use DiscoveryLink DDL statements to create new data collections at relational data sources. Another feature allows certain queries to be answered using pre-materialized automatic summary tables stored by DiscoveryLink, with little or no access to the data sources themselves. Joins, subqueries, table expressions, aggregation, statistical functions, and many other SQL constructs are supported against data, whether the data is locally stored or retrieved from remote data sources.
11.2.1 Query Optimization
During the planning process, the DiscoveryLink server takes into account the query processing power of each data source. As it identifies query fragments to be performed at a data source, it must ensure that the fragments are executable by that source. If a fragment cannot be performed by the source, the optimizer builds a plan to compensate for the missing function by doing that piece of work in the DiscoveryLink server. For example, if the data source does not do joins, but it is necessary to join together data from two nicknames at that source, the data will be retrieved from both nicknames (typically after restricting it with any predicates the source can apply), and then joined by DiscoveryLink.
The DiscoveryLink server has two ways of obtaining information about query processing power. Wrappers provided by IBM for relational data sources (and for other sources that are similar to a relational source in function) provide a server attributes table (SAT). The SAT contains a long list of parameters that are set to appropriate values by the wrapper. For example, if the parameter PUSHDOWN is set to “N”, DiscoveryLink will not request that the data source perform query fragments more complex than:
![]()
Note: In this chapter, SQL is used as a concise way of expressing the work to be done by a remote data source. This work is actually represented internally by various data structures for efficient data processing.
If PUSHDOWN is set to ‘Y’, more complex requests may be generated, depending on the nature of the query and the values of other SAT parameters. For example, if the value of the BASIC_PRED parameter in the SAT is ‘Y’, requests may include predicates such as:
![]()
The parameter MAX_TABS is used to indicate a data source’s ability to perform joins. If it is set to 1, no joins are supported. Otherwise MAX_TABS indicates the maximum number of nicknames that can appear in a single FROM clause of the query fragment to be sent to the data source.
Information about the cost of query processing by a data source is supplied to the DiscoveryLink optimizer in a similar way, using a fixed set of parameters such as CPU_RATIO, which is the relative speed of the data source’s processor relative to the one hosting the DiscoveryLink server. Additional parameters, such as average number of instructions per invocation and average number of Input/Output (I/O) operations per invocation, can be provided for data source functions defined to DiscoveryLink with function mappings, as can statistics about tables defined as nicknames. Once defined, these parameters and statistics can be easily updated whenever necessary.
This approach has proven satisfactory for describing the query processing capabilities and costs of the relational database engines supported by DiscoveryLink; although even for these superficially similar sources, a large set (hundreds) of parameters is needed. However, it is difficult to extend this approach to more idiosyncratic data sources. Web servers, for example, may be able to supply many pieces of information about some entity, but frequently they will only allow certain attributes to be used as search criteria. This sort of restriction is difficult to express using a fixed set of parameters. Similarly, the cost of executing a query fragment at a data source may not be easily expressed in terms of fixed parameters, if, for example, the cost depends on the value of an argument to a function. For instance, a BLAST function asked to do a BLASTp comparison against a moderate amount of data will return in seconds, whereas if it is asked to do a tBLASTn comparison against a large dataset, it may need hours.
The solution, validated in the Garlic prototype, is to involve the wrappers directly in planning of individual queries. Instead of attempting to model the behavior of a data source using a fixed set of parameters with statically determined values, the DiscoveryLink server will generate requests for the wrapper to process specific query fragments. In return, the wrapper will produce one or more wrapper plans, each describing a specific portion of the fragment that can be processed, along with an estimate for the cost of computing the result and its estimated size.
11.2.2 An Example
Voltage-sensitive calcium channel proteins mediate the entry of calcium ions into cells and are involved in such processes as neurotransmitter release. They respond to electric changes, which are a prominent feature of the neural system. The discovery of a novel gene that codes for a calcium channel protein would potentially be of great interest to pharmaceutical researchers seeking new drug targets for a neuropsychological disease. A popular method of novel gene discovery is to search Expressed Sequence Tag (EST) databases for (expressed) sequences similar to known genes or proteins. For example, a scientist with access to the data sources just described might like to see the results of the following query:
“Return accession numbers and definitions of EST sequences that are similar (60% identical over 50 amino acids) to calcium channel sequences in the protein data source that reference papers published since 1995 mentioning ‘brain’.”
The hsp_info column holds a condensed form of the equivalent data in the XML specification for BLAST provided by the National Center for Biotechnology Information. But to answer the above query, one needs direct access to the percentage of identities within the hsp alignment and the length of that alignment. Assume that two user-defined functions are defined to extract this information from the hsp_info string:
![]()
and
![]()
Then, this request can be expressed as a single SQL statement that combines data from all three data sources:

Many possible evaluation plans exist for this query. One plan is shown in Figure 11.3. In this figure, each box represents an operator. The leaves represent actions at a data source. Because DiscoveryLink does not model the details of those actions, each action is modeled as a single operator, even if it might involve a series of operators at the source. (For relational sources, DiscoveryLink does in fact model the individual operators, but to simplify the figures, details are omitted. Thus the join of Proteins and Prot-Pubs is modeled in the figures as a single operator.) For non-relational sources, DiscoveryLink would not know whether a logical join action was an actual join or whether, in fact, the data was stored in a nested, pre-joined format. Nor does DiscoveryLink know whether the data are scanned and then predicates applied, or there is an indexed access, and so on. Instead, DiscoveryLink keeps track of the work that has been done by recording the properties of each operator. The properties include the set of columns available (C), the set of tables accessed (T), and the set of predicates applied (P), as shown in Figure 11.3. Non-leaf nodes represent individual operations at the DiscoveryLink server. The optimizer models these local operations separately.
This plan first accesses the protein data source, retrieving protein names and sequences and corresponding publication identifiers, for all proteins that serve as calcium channels. This information is returned to DiscoveryLink, where the bindjoin operator sends the publication references to the publications source one at a time. At the publications source, these publication identifiers are used to find relevant publications, and those publications are further checked for compliance with the query restrictions on keyword and pub_date. For those publications that pass all the tests, the identifier is returned to DiscoveryLink. There, the second bindjoin operator sends the sequence for any surviving proteins to BLAST, where they are compared against gbest, and the results are returned to DiscoveryLink where each hsp_info is analyzed to see if the sequence is sufficiently similar.
A second, superficially similar plan is shown in Figure 11.4. In this plan, the publications with appropriate dates and keywords are sent to DiscoveryLink, where a hash table is built. The data from the protein data source are also sent to DiscoveryLink and used to probe the hash table. Matches are passed to the bindjoin operator, which BLASTs the sequences against gbest, then returns them to DiscoveryLink to check the quality of the match.
It is not obvious which plan is best. The first plan results in one query of the protein database, but many queries (one for each qualified protein) of the publications database. The second plan only queries each of these sources once, but potentially returns many publication entries for proteins that will not qualify.
Either of these plans is likely to be better than the one shown in Figure 11.5. In this plan, the protein data is extracted first and all calcium channel proteins are BLASTed against gbest, regardless of what publications they reference. DiscoveryLink then filters the sequences using the similarity criterion, and the remaining proteins are passed to the nested loop join operator. This join compares each protein’s referenced publications with a temporary table created by storing in DiscoveryLink those recent publications that discuss the brain. This plan could only win if there were very few recent publications with brain as a keyword (so the cost of the query to make the temporary table is small), and yet virtually every calcium channel protein in the protein database referenced at least one of them (so there is no benefit to doing the join of proteins with publications early). While that is unlikely for this example, if there were a more restrictive set of predicates (e.g., a recently discovered protein of interest and papers within the last two months), this plan could, in fact, be a sensible one.
11.2.3 Determining Costs
Accurately determining the cost of the various possible plans for this or any query is difficult for several reasons. One challenge is estimating the cost of evaluating the wrapper actions. For example, the DiscoveryLink engine has no notion of what must actually be done to find similar sequences or how the costs will vary depending on the input parameters (the bound columns). For BLAST, the actual algorithm used can change the costs dramatically, as can the data set being searched. As a second challenge, the query processor has no way to estimate the number of results that may be returned by the data sources. While the wrapper could, perhaps, provide some statistics to DiscoveryLink, purely relational statistics may not be sufficient. For example, cardinalities, as well as costs, for search engines like BLAST may vary depending on the inputs. A third challenge is to estimate the cost of the functions in the query. The costing parameters maintained by relational wrappers in DiscoveryLink for a function implemented by a data source include a cost for the initial invocation and a per-row cost for each additional invocation. However, the only way to take the value of a function argument into account is through a cost adjustment based on the size of the argument value in bytes. While this may be acceptable for simple functions like percent_identity and align_length, it is unlikely, in general, to give accurate results. For example, if contains actually has to search in different ways depending on the type of the column passed as an argument (e.g., a simple scan for keyword but an index lookup for the paper itself), the cost parameters must be set to reflect some amalgamation of all the search techniques. A simple case statement, easily written by the wrapper provider, could model the differences and allow more sensible choices of plans. While the costs of powerful functions in some cases can be hard to predict, many vendors do, in fact, know quite a bit about the costs of their functions. They often model costs themselves to improve their systems’ performance.
The challenges of accurately estimating costs are met by letting the wrapper examine possible plan fragments to provide information about what the data source can do and how much it will cost. Consider our example query once again. During the first phase of optimization, when single-table access plans are being considered, the publications database will receive the following fragment for consideration (again, query fragments are represented in SQL; the actual wrapper interface uses an equivalent data structure that does not require parsing by the wrapper).
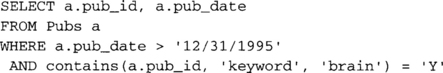
Assume that, in a single operation, the publications database can apply either the predicate on publication date or the contains predicate, but not both. Many Web sites can handle only a single predicate at a time or only restricted combinations. (Note: In the previous illustrative plans, it was assumed that the publications database could do both. Either assumption might be true. This one is adopted here to illustrate the point.) Further assume that it is possible to invoke the contains function separately later (this is like asking a new, very restrictive query of the Web site). Many Web sites do allow such follow-on queries to retrieve additional information about an object or do some more complex computation. The wrapper might return two wrapper plans for this fragment. The first would indicate that the data source could perform the following portion of the fragment:

with an estimated execution cost of 3.2 seconds and an estimated result size of 500 publications (in reality, of course, the result size would be much bigger). To estimate the total cost of the query fragment using this wrapper plan, the DiscoveryLink optimizer would add to the cost for the wrapper plan the cost of invoking the contains function on each of the 500 publications returned. If each invocation costs a second (because of the high overhead of going out to the World Wide Web), the total cost of this portion of the query, using this plan, would be 503.2 seconds.
The second wrapper plan would indicate that the data source could perform the following portion of the fragment:

with an estimated execution cost of 18 seconds and an estimated result size of 1000 publications (i.e., entries for all the publications in the database with the keyword brain). To compute the total cost in this case, the optimizer would augment the cost for the wrapper plan with the cost of using the DiscoveryLink engine to apply the predicate on publication date to each of the 1000 publications. If filtering one publication takes a 1/100 of a second, the total cost for this portion of the query, using this plan, would be 28 seconds—a clear winner.
Wrappers participate in query planning in the same way during the join enumeration portion of optimization. In the example, the wrapper might be asked to consider the following query fragment:

This is essentially a single-table access, but the third predicate would not be considered during single-table access planning because the value being compared to pub_id comes from a different table. For each pub_id produced by the rest of the query (represented above by the host variable :H0), the publications database is asked to find the important properties of the corresponding publication, if it matches the other criteria. As before, the wrapper would return one or more plans and indicate in each one which of the predicates would be evaluated.
Only a few of the plans that DiscoveryLink would consider in optimizing this query were shown. The goal was not to give an exhaustive list of alternatives, but rather to illustrate the process. As well, the chapter has demonstrated the critical role an optimizer plays for complex queries. It is neither obvious nor intuitive which plan will ultimately be the best; the answer depends on many factors including data volumes, data distributions, the speeds of different processors and network connections, and so on. Simple heuristics generally cannot arrive at the right answer. Only a cost-based process with input on specific data source characteristics can hope to choose the right plans for the vast array of possible queries.
As a wrapper may be asked to consider many query fragments during the planning of a single query, it is important that communication with the wrapper be efficient. This is achieved easily in DiscoveryLink because the shared library that contains a wrapper’s query planning code is loaded on demand into the address space of the DiscoveryLink server process handling the query. The overhead for communicating with a wrapper is, therefore, merely the cost of a local procedure call.
This approach to query planning has many benefits. It is both simple and extremely flexible. Instead of using an ever-expanding set of parameters to invest the DiscoveryLink server with detailed knowledge of each data source’s capabilities, this knowledge resides where it falls more naturally, in the wrapper for the source in question. This allows to exploit the special functionality of the underlying source, as was done for the BLAST server (by modeling the search algorithm as a virtual table) and the publications source (using a template function). The wrapper only responds to specific requests in the context of a specific query. As the previous examples have shown, sources that only support searches on the values of certain fields or on combinations of fields are easily accommodated. In a similar way, one can accommodate sources that can only sort results under certain circumstances or can only perform certain computations in combination with others. Because a wrapper needs only to respond to a request with a single plan, or in some cases no plan at all, it is possible to start with a simple wrapper that evolves to reflect more of the underlying data source’s query processing power.
This approach to query planning need not place too much of a burden on the wrapper writer, either. In a paper presented at the annual conference on very large databases [27], Roth et al. showed that it is possible to provide a simple default cost model and costing functions along with a utility to gather and update all necessary cost parameters. The default model did an excellent job of modeling simple data sources and did a good job predicting costs, even for sources that could apply quite complex predicates. This same paper further showed that even an approximate cost model dramatically improved the choice of plans over no information or fixed default values [27]. Therefore, it is believed that this method of query planning is not only viable, but necessary. With this advanced system for optimization, DiscoveryLink has the extensibility, flexibility, and performance required to meet the needs of life sciences applications.
11.3 EASE OF USE, SCALABILITY, AND PERFORMANCE
DiscoveryLink provides a flexible platform for building life sciences applications. It is not intended for the scientist, but rather for the application programmer, an IT worker, or a vendor who creates the tools that the actual scientists will use. While it provides only a simple user interface, it supports multiple programming interfaces, including such de facto industry standards as ODBC and JDBC. It, therefore, can be used with any commercially available tool that supports these interfaces, including popular query builders, such as those by Brio or Cognos, application-building frameworks such as VisualAge from IBM, or industry-specific applications including LabBook, Spotfire, and so on. Alternatively, in-house applications can be developed that meet the needs of specific organizations. IBM has a number of business partners who are including DiscoveryLink in their offerings to create a more complete scientific workbench for their customers.
Database Administrators will also be users of DiscoveryLink. For these, DiscoveryLink has a Graphical User Interface (GUI) to help with the registration process. Yet life sciences applications require more support than this. Complete integration also requires the development of tools to bridge between different models of data. The life sciences research community is not a homogeneous one. Different groups use different terms for the same concept or describe different concepts similarly. Semantic mappings must be created, and applications for particular communities must be developed. DiscoveryLink provides features that help with these tasks, but it does not solve either. For example, views can help with the problems of semantic integration by hiding mappings from one data representation to another, but the views still must be created manually by the DBAs.
Another characteristic of life sciences data and research environments is frequent change, both in the amounts of data and in the schemas in which data is stored (causing more work for DBAs). Further, new sources of information are always appearing as new technologies and informatics companies evolve. In such an environment, flexibility is essential. DiscoveryLink’s powerful query processor and non-procedural SQL interface protect applications (to the extent possible) from changes in the underlying data source via the principle of logical data independence. New sources of information require a new server definition, however, and perhaps a new wrapper, and may also require adjusting view definitions to reference their data. Changes in a data source’s interfaces often can be hidden from the application by modifying the translation portion of the wrapper or installing a new wrapper with the new version of the source. The query processing technology is built to handle complex queries and to scale to terabytes of data. Thus, the database middleware concept itself allows DiscoveryLink to deal with the changes in this environment, but it puts a burden on the DBA to administer these changes.
Wrapper writers are a third group of users. The wrapper architecture has been designed for extensibility. Only a small number of functions need to be written to create a working wrapper. Simple sources can be wrapped quickly, in a week or two; more complex sources may require from a few weeks to a few months to completely model, but even for these a working wrapper, perhaps with limited functionality, can be completed quickly. Template code for each part of the wrapper and default cost modeling code are provided for wrapper writers. Wrappers are built to enable as much sharing of code as possible, so that one wrapper can be written to handle multiple versions of a data source, and so that wrappers for similar sources can build on existing wrappers. The ability to separate schema information from wrapper code means that changes in the schema of a data source require no code changes in the wrappers. The addition of a new data source requires no change to any existing wrappers. Thus, the wrappers also help the system adapt to the many changes possible in the environment, and the wrapper architecture eases the wrapper writer’s task.
Scalability is a fundamental goal of DiscoveryLink. There is no a priori limit to the number of different sources it can handle, because sources are independent and consume little in the way of system resources when not in use. (Wrapper code is loaded dynamically; when not in use, the only trace of the source is a set of catalog entries.) There may be limitations in practice if many sources of different types are used at the same time, depending on how much memory is available. This is akin to the limits on query complexity in relational database management systems today, which are not typically hit until several hundred tables are used in the same query. Because DiscoveryLink is built on robust and scalable relational database technology, there should also be no a priori limit on the amount of data the system can handle. Because the data are left in the native stores until needed, they can still be updated by directly modifying those stores. (That is, updates do not need to go through DiscoveryLink, though it may be convenient to do so for relational data sources.) In this case, the update rate is only limited by the update rate of the data sources; DiscoveryLink is not a bottleneck.
As with all database management systems, DiscoveryLink needs to be able to handle complex queries over large volumes of data swiftly and efficiently. For DiscoveryLink, this task is further complicated by the fact that much, if not all, of the data resides in other data sources, which may be distributed over a wide geographic area. Query optimization, which is described in this chapter, is the main tool DiscoveryLink uses to ensure good performance. There are other aspects of the system that also help. For example, before optimization, the query is passed to a rewrite engine. This engine applies a variety of transformations that can greatly improve the ultimate performance. Transforms can, for example, eliminate unnecessary operations such as sorts or even joins. Others can derive new predicates that restrict operations or allow the use of a different access path, again enhancing performance. In addition to query rewrite, wrappers are carefully tuned to use the most efficient programming interfaces provided by the source (e.g., taking advantage of bulk reads and writes to efficiently transport data between sources). Additional constructs such as automatic summary tables (materialized views over local and/or remote data that can be automatically substituted into a query to save remote data access and re-computation) provide a simple form of caching.
How well does the system perform? There are no benchmarks yet for this style of federated data access, and IBM experience to date is limited to a few customers and some experiments in IBM’s lab. But, some statements can be made. For example, it is known that DiscoveryLink adds little if any overhead. A simple experiment compares queries that can be run against a single source with the same query submitted against a DiscoveryLink nickname for that source. In most cases, the native performance and performance via DiscoveryLink are indistinguishable [8]. In a few cases, due either to the sophisticated rewrite engine or just the addition of more hardware power, performance using this three-tiered approach (client→Discovery Link→source) is better than performance using the source directly (client→source). This has been borne out by repeated experiments on both customer and standard TPC-H workloads. For queries that involve data from multiple sources, it is harder to make broad claims, as there is no clear standard for comparison. The overall experience so far shows that performance depends heavily on the complexity of the query and the amounts of data that must be transported to complete the query. Overall, performance seems to be meeting customers’ needs; that is, it is normally good enough that they do not shy away from distributed queries and often are not even aware of the distribution. There are areas for improvement, however, including better exploitation of parallelism when available and some form of automated caching.
11.4 CONCLUSIONS
This chapter described IBM’s DiscoveryLink offering. DiscoveryLink allows users to query data that may be physically stored in many disparate, specialized data stores as if that data were all co-located in a single virtual database. Queries against this data may exploit all of the power of SQL, regardless of how much or how little SQL function the data sources provide. In addition, queries may employ any additional functionality provided by individual data stores, allowing users the best of both the SQL and the specialized data source worlds. A sophisticated query optimization facility ensures that the query is executed as efficiently as possible. The interfaces, performance, and scalability of DiscoveryLink were also discussed.
DiscoveryLink is a new offering, but it is based on a fusion of well-tested technologies from DB2 Universal Database (UDB), DB2 DataJoiner, and the Garlic research project. Both DB2 UDB (originally DB2 Client/Server [C/S]) and DB2 DataJoiner have been available as products since the early 1990s, and they have been used by thousands of customers in the past decade. The Garlic project began in 1994, and much of its technology was developed as the result of joint studies with customers, including an early study with Merck Research Laboratories. DiscoveryLink’s extensible wrapper architecture and the interactions between wrapper and optimizer during query planning derive from Garlic. As part of Garlic, wrappers were successfully built and queried for a diverse set of data sources, including two relational database systems (DB2 and Oracle), a patent server stored in Lotus Notes, searchable sites on the World Wide Web (including a database of business listings and a hotel guide), and specialized search engines for collections of images, chemical structures, and text.
Currently, IBM is working on building a portfolio of wrappers specific to the life sciences industry. In addition to key relational data sources such as Oracle and Microsoft’s SQL Server, wrappers are available for application sources such as BLAST and general sources of interest to the industry such as Microsoft Excel, flat files, Documentum for text management, and XML. IBM is also working with key industry vendors to wrap the data sources they supply. This will provide access to many key biological and chemical sources. While wrappers will be created as quickly as possible, it is anticipated that most installations will require one or more new wrappers to be created because of the sheer number of data sources that exist and the fact that many potential users have their own proprietary sources as well. Hence, a set of tools is being developed for writing wrappers and training a staff of wrapper writers who will be able to build new wrappers as part of the DiscoveryLink software and services offering model. As DiscoveryLink supports the SQL/MED standard [25] for accessing external data sources, those who would rather create their own wrappers (customers, universities, and business partners) may do so, too. Hopefully, in this way a rich set of wrappers will quickly become available for use with DiscoveryLink.
From the preceding pages, hopefully it is clear that DiscoveryLink plays an essential role in integrating life science data. DiscoveryLink provides the plumbing, or infrastructure, that enables data to be brought together, synthesized, and transformed. This plumbing provides a high-level interface, a virtual database against which sophisticated queries can be posed and from which results are returned with excellent performance. It allows querying of heterogeneous collections of data from diverse data sources without regard to where they are stored or how they are accessed.
While not a complete solution to all heterogeneous data source woes, DiscoveryLink is well suited to the life sciences environment. It serves as a platform for data integration, allowing complex cross-source queries and optimizing them for high performance. In addition, several of its features can help in the resolution of semantic discrepancies by providing mechanisms DBAs can use to bridge the gaps between data representations. Finally, the high-level SQL interface and the flexibility and careful design of the wrapper architecture make it easy to accommodate the many types of change prevalent in this environment.
Of course, there are plenty of areas in which further research is needed. For the query engine, key topics are the exploitation of parallelism to enhance performance and richer support for modeling of object features in foreign data sources. There is also a need for additional tools and facilities that enhance the basic DiscoveryLink offering. Some preliminary work was done on a system for data annotation that provides a rich model of annotations, while exploiting the DiscoveryLink engine to allow querying of annotations and data separately and in conjunction. A tool is also being built to help users create mappings between source data and a target, integrated schema [28, 29] to ease the burden of view definition and reconciliation of schemas and data that plagues today’s system administrators. Hopefully, as DiscoveryLink matures it will serve as a basis for more advanced solutions that will distill information from the oceans of data in which life sciences researchers are currently drowning, for the advancement of human health and for basic scientific understanding.
REFERENCES
[1] Altschul, S., Gish, W., Miller, W., et al. Basic Local Alignment Search Tool. Journal of Molecular Biology. 1990;215(no. 3):403–410.
[2] Falquet, L., Pagni, M., Bucher, P., et al. The PROSITE Database Its Status in 2002. Nucleic Acids Research. 2002;30(no. 1):235–238.
[3] [see comments] Birney, E., Durbin, R. Using Gene Wise in the Drosophilia Annotation Experiment. Genome Research. 2000;10(no. 4):547–548.
[4] Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., et al. GenBank. Nucleic Acids Research. 2002;30(no. 1):17–20.
[5] Bairoch, A., Apweiler, R. The SWISS_PROT Protein Sequence Database and Its Supplement TrEMBL in 2000. Nucleic Acids Research. 2000;28(no. 1):45–48.
[6] Wheeler, D.L., Church, D.M., Lash, A.E., et al. Database Resources of the National Center for Biotechnology Information: 2002 Update. Nucleic Acids Research. 2002;20(no. 1):13–16.
[7] Ringwald, M., Epping, J.T., Begley, D.A., et al. The Mouse Gene Expression Database (GXD). Nucleic Acids Research. 2001;29(no. 1):98–101.
[8] February Haas, L.M., Schwarz, P.M., Kodali, P., et al. DiscoveryLink: A System for Integrated Access to Life Sciences Data Sources. IBM Systems Journal. 2001;40(no. 2):489–511.
[9] Gupta, P., Lin, E.T. Datajoiner: A Practical Approach to Multi-Database Access. In: Proceedings of the International IEEE Conference on Parallel and Distributed Information Systems. Los Alamitos, CA: IEEE Computer Society; 1994:264.
[10] Haas, L.M., Kossmann, D., Wimmers, E.L., et al. Optimizing Queries Across Diverse Data Sources. In: Proceedings of the Conference on Very Large Databases (VLDB). San Francisco: Morgan Kaufmann; 1997:276–285.
[11] Roth, M.T., Schwarz, P.M. Don’t Scrap It, Wrap It! A Wrapper Architecture for Legacy Data Sources. In: Proceedings of the Conference on Very Large Data Bases (VLDB). San Francisco: Morgan Kaufmann; 1997:266–275.
[12] January Davidson, S., Overton, C., Tannen, V., et al. BioKleisli: A Digital Library for Biomedical Researchers. International Journal of Digital Libraries. 1997;1(no. 1):36–53.
[13] Etzold, T., Argos, P. SRS: An Indexing and Retrieval Tool for Flat File Data Libraries. Computer Applications in the Biosciences. 1993;9(no. 1):49–57.
[14] Carter, P., Coupaye, T., Kreil, D., et al. SRS: Analyzing and Using Data from Heterogeneous Textual Databanks. In: Letovsky S., ed. Bioinformatics: Databases and Systems. Boston: Kluwer Academic, 1998.
[15] Chen, I-M.A., Kosky, A.S., Markowitz, V.M., et al. Constructing and Maintaining Scientific Database Views in the Framework of the Object-Protocol Model. In: Proceedings of the Ninth International Conference on Scientific and Statistical Database Management. Los Alamitos, CA: IEEE Computer Society; 1997:237–248.
[16] Stevens, R., Goble, C., Paton, N.W., et al. Complex Query Formulation Over Diverse Information Sources in TAMBIS. In: Lacroix Z., Critchlow T., eds. Bioinformatics: Managing Scientific Data. San Francisco: Morgan Kaufmann; 2004:189–223.
[17] Eckman, B.A., Kosky, A.S., Laroco, L.A., Jr. Extending Traditional Query-Based Integration Approaches for Functional Characterization of Post-Genomic Data. Bioinformatics. 2001;17(no. 7):587–601.
[18] Papakonstantinou, Y., Garcia-Molina, H., Widom, J. Object Exchange Across Heterogeneous Information Sources. In: Proceedings of the IEEE Conference on Data Engineering. Los Alamitos, CA: IEEE Computer Society; 1995:251–260.
[19] Tomasic, A., Raschid, L., Valduriez, P. Scaling Heterogeneous Databases and the Design of DISCO. In: Proceedings of International Conference on Distributed Computing Systems (ICDCS). Los Alamitos, CA: IEEE Computer Society; 1996:449–457.
[20] Shan, M-C., Ahmed, R., Davis, J., et al. Pegasus: A Heterogeneous Information Management System. In: Kim W., ed. Modern Database Systems. Reading, MA: Addison-Wesley; 1995:664–682.
[21] Liu, L., Pu, C. The Distributed Interoperable Object Model and its Application to Large-Scale Interoperable Database Systems. In: Proceedings of the ACM International Conference on Information and Knowledge Management. New York: Association for Computing Machinery; 1995:105–112.
[22] Adali, S., Candan, K., Papakonstantinou, Y., et al. Query Caching and Optimization in Distributed Mediator Systems. In: Proceedings of the ACM SIGMOD Conference on Management of Data. New York: Association for Computing Machinery; 1996:137–148.
[23] International Organization for Standardization. Information Technology — Database Languages—SQL—Part 3: Call Level Interface (SQL/CLI). In: 15O/IEC 9075-3. Geneva, Switzerland: International Organization for Standardization; 1999.
[24] International Organization for Standardization. Information Technology — Database Language—SQL—Part 2: Foundation (SQL/Foundation). In: ISO/IEC 9075-2. Geneva, Switzerland: International Organization for Standardization; 1999.
[25] International Organization for Standardization. Information Technology Database Languages—SQL—Part 9: Management of External Data (SQL/MED). In: ISO/IEC 9075-9. Geneva, Switzerland: International Organization for Standardization; 2000.
[26] Version 7.2 FP 5 IBM, IBM DB2 Life Sciences Data Connect Planning, Installation and Configuration Guide. White Plains, NY: IBM; 2001. 3.ibm.com/software/data/db2/lifesciencesdataconnect/db21s
[27] Roth, M.T., Ozean, F., Haas, L.M. Cost Models Do Matter: Providing Cost Information for Diverse Data Sources in a Federated System. In: Proceedings of the Conference on Very Large Data Bases (VLDB). San Francisco: Morgan Kaufmann; 1999:559–610.
[28] Haas, L.M., Miller, R.J., Niswonger, B., et al. Transforming Heterogeneous Data with Database Middleware: Beyond Integration. IEEE Data Engineering Bulletin. 1999;22(no. 1):31–36.
[29] Miller, R.J., Haas, L.M., Hernandez, M. Schema Mapping as Query Discovery. In: Proceedings of the Conference on Very Large Data Bases (VLDB). San Francisco: Morgan Kaufmann; 2000:77–88.





