A Practitioner’s Guide to Data Management and Data Integration in Bioinformatics
3.1 INTRODUCTION
Integration of a large and widely diverse set of data sources and analytical methods is needed to carry out bioinformatics investigations such as identifying and characterizing regions of functional interest in genomic sequence, inferring biological networks, and identifying patient sub-populations with specific beneficial or toxic reactions to therapeutic agents. A variety of integration tools are available, both in the academic and the commercial sectors, each with its own particular strengths and weaknesses. Choosing the right tools for the task is critical to the success of any data integration endeavor. But the wide variety of available data sources, integration approaches, and vendors makes it difficult for users to think clearly about their needs and to identify the best means of satisfying them. This chapter introduces use cases for biological data integration and translates them into technical challenges. It introduces terminology and provides an overview of the landscape of integration solutions, including many that are detailed in other chapters of this book, along with a means of categorizing and understanding individual approaches and their strengths and weaknesses.
This chapter is written from the point of view of a bioinformatician practicing database integration, with the hope that it will be useful for a wide variety of readers, from biologists who are unfamiliar with database concepts to more computationally experienced bioinformaticians. A basic familiarity with common biological data sources and analysis algorithms is assumed throughout.
The chapter is organized as follows. Section 3.2 introduces traditional database terms and concepts. Those already familiar with these concepts may want to skim that section and begin reading at Section 3.3, which introduces multiple dimensions to integration, thus intermediate terminology. Section 3.4 presents various use cases for integration solutions. Strengths and weaknesses of integration approaches are given in Section 3.5. Section 3.6 is devoted to tough integration problems. Therefore, computer scientists and information technologists may benefit from the advanced problems evoked in Section 3.6.
The goal of this chapter is to convey a basic understanding of the variety of data management problems and needs in bioinformatics; an understanding of the variety of integration strategies currently available, and their strengths and weaknesses; an appreciation of some difficult challenges in the integration field; and the ability to evaluate existing or new integration approaches according to six general categories or dimensions. Armed with this knowledge, practitioners will be well prepared to identify the tools that are best suited to meet their individual needs.
3.2 DATA MANAGEMENT IN BIOINFORMATICS
Data is arguably the most important commodity in science, and its management is of critical importance in bioinformatics. One introductory textbook defines bioinformatics as “the science of creating and managing biological databases to keep track of, and eventually simulate, the complexity of living organisms” [1]. If the central task of bioinformatics is the computational analysis of biological sequences, structures, and relationships, it is crucial that biological sequence and all associated data be accurately captured, annotated, and maintained, even in the face of rapid growth and frequent updates. It is also critical to be able to retrieve data of interest in a timely manner and to define and retrieve data of interest precisely enough to separate effectively its signal from the distracting noise of irrelevant or insignificant data.
3.2.1 Data Management Basics
To begin the discussion of data management in bioinformatics, basic terms and concepts will be introduced by means of use cases, examples or scenarios of familiar data management activities. The term database will be used both as “a collection of data managed by a database management system” (DBMS) and, more generally, when concepts of data representation are presented, regardless of how the data is managed or stored. Otherwise, the term data collection or data source will be used for collections of data not managed by a DBMS. For a more detailed explanation of basic data management than is possible in this chapter, see Ullman and Widom’s A First Course in Database Systems [2].
Use Case: A Simple Curated Gene Data Source
Consider a simple collection of data about known and predicted human genes in a chromosomal region that has been identified as likely to be related to a genetic predisposition for a disease under investigation. The properties stored for each gene are as follows:
![]() GenBank accession number (accnum) [3]
GenBank accession number (accnum) [3]
![]() Aliases in other data sources (e.g., Swiss-Prot accession number) [4]
Aliases in other data sources (e.g., Swiss-Prot accession number) [4]
![]() Protein families database (Pfam) classification [5]
Protein families database (Pfam) classification [5]
![]() Gene Ontology (GO) annotation [6]
Gene Ontology (GO) annotation [6]
![]() Has expression results? (Are there expression results for this gene?)
Has expression results? (Are there expression results for this gene?)
![]() Has Single Nucleotide Polymorphisms (SNPs)? (Are there known SNPs for this gene?)
Has Single Nucleotide Polymorphisms (SNPs)? (Are there known SNPs for this gene?)
The complement of properties stored in a database, along with the relationships among them, is called the database’s schema. Individual properties, GenBank accession number, are attributes. Attributes can be single-valued, like peptide sequence, or multi-valued, like aliases. Attributes can be atomic, like peptide sequence, which is a simple character string, or nested, like aliases, which themselves have structure (data source + identifier).
Data accuracy is critically important in scientific data management. Single attributes or groups of attributes must satisfy certain rules or constraints for the data to be valid and useful. When entering data into the database, or populating it, care must be taken to ensure that these constraints are met. Examples of constraints in the simple gene data source are:
![]() The chromosomal location of each gene must lie within the original region of interest.
The chromosomal location of each gene must lie within the original region of interest.
![]() The CDS and peptide sequences must contain only valid nucleotide and amino acid symbols, respectively.
The CDS and peptide sequences must contain only valid nucleotide and amino acid symbols, respectively.
![]() The CDS sequence must have no internal in-frame stop codons (which would terminate translation prematurely).
The CDS sequence must have no internal in-frame stop codons (which would terminate translation prematurely).
![]() The peptide sequence must be a valid translation of the CDS sequence.
The peptide sequence must be a valid translation of the CDS sequence.
![]() The Pfam classification must be a valid identifier in the Pfam data source.
The Pfam classification must be a valid identifier in the Pfam data source.
This simple gene data collection is subject to continual curation, in which new data is inserted, old data is updated, and erroneous data is deleted. A user might make changes to existing entries as more information becomes known, such as more accurate sequence or exon boundaries of a predicted gene, refined GO classification, SNPs discovered, or expression results obtained. A user might also make changes to the source’s schema, such as adding new attributes like mouse orthologues, or links to LocusLink, RefSeq [7], or KEGG pathways [8]. New linkage studies may result in a widening or narrowing of the chromosomal region of interest, requiring a re-evaluation of which genes are valid members of the collection and the addition or deletion of genes. Finally, multiple curators may be working on the data collection simultaneously. Care must be taken that an individual curator’s changes are completed before a second curator’s changes are applied, lest inconsistencies result (e.g., if one curator changes the CDS sequence and the other changes the peptide sequence so they are no longer in the correct translation relationship to one another). The requirement for correctly handling multiple, simultaneous curators’ activities is called multi-user concurrency.
Databases are only useful, of course, if data of interest can be retrieved from them when needed. In a small database, a user might simply need to retrieve all the attributes at once in a report. More often, however, users wish to retrieve subsets of a database by specifying conditions, or search predicates, that the data retrieved should meet. Examples of queries from the curated gene data collection described previously are: “Retrieve the gene whose GenBank accession number is AA123456”; “retrieve only genes that have expression results”; “retrieve only genes that contain in their description the words ‘serotonin receptor.’” Search predicates may be combined using logical AND and OR operators to produce more complex conditions; for example, in the query “Retrieve genes which were entered since 09/01/2002 and lie in a specified sub-region of the chromosomal region of interest,” the conjunction of the two search predicates will be expressed by an operator AND.
Use Case: Retrieving Genes and Associated Expression Results
Along with the simple curated gene data collection, a user may wish to view expression data on the genes that have been gathered through microarray experiments. For example, an expression data source might permit the retrieval of genes that show equal to or greater than two-fold difference in expression intensities between ribonucleic acid (RNA) isolated from normal and diseased tissues. To retrieve all genes with known SNPs with at least two-fold differential expression between normal and diseased tissue, search predicates would need to be applied to each of the two data sources. The result would be the genes that satisfy both of the conditions.
There are many different but equivalent methods of retrieving the genes that satisfy both of these predicates, and an important task of a database system is to identify and execute the most efficient of these alternate methods. For example, the system could first find all genes with SNPs from among the curated genes, and then check the expression values for each of them one by one in the expression data source. Alternatively, the system could find all genes in the expression data source with two-fold expression in normal versus diseased tissue, then find all genes in the curated data source that have SNPs, and finally merge the two lists, retaining only the genes that appear in both lists. Typically, methods differ significantly in their speed due to such factors as the varying speeds of the two databases, the volume of data retrieved, the specificity of some predicates, the lack of specificity of others, and the order in which predicates are satisfied. They may also differ in their usage of computer system resources such as central processing unit (CPU) or disk. Depending on individual needs, the execution cost may be defined either as execution time or resource usage (see Chapter 13). The process of estimating costs of various alternative data retrieval strategies and identifying the lowest one among them is known as cost-based query optimization.
3.2.2 Two Popular Data Management Strategies and Their Limitations
Two approaches that have commonly been used to manage and distribute data in bioinformatics are spreadsheets and semi-structured text files.
Spreadsheets
Spreadsheets are easy to use and handy for individual researchers to browse their data quickly, perform simple arithmetic operations, and distribute them to collaborators. The cell-based organization of a spreadsheet enables the structuring of data into separate items, by which the spreadsheet may then be sorted. The Microsoft Excel spreadsheet software [9] provides handy data entry features for replicating values in multiple cells, populating a sequence of rows with a sequence of integer identifiers, and entering values into a cell that have appeared in the same column previously.
A disadvantage of spreadsheets, at least as they are typically used, is that very little data validation is performed when data is entered. It is certainly possible, by programming in Microsoft Visual Basic or using advanced Excel features, to perform constraint checking such as verifying that data values have been taken from an approved list of values or controlled vocabulary, that numeric data fall in the correct range, or that a specific cell has not been left blank; but in practice this is not often done. Furthermore, while advanced features exist to address this problem, in practice spreadsheets typically include a great deal of repeated or redundant data. For example, a spreadsheet of gene expression data might include the following information, repeated for each tissue sample against which the gene was tested: GenBank accession number, gene name, gene description, LocusLink Locus ID, and UniGene Cluster ID. If an error should be found in any of these redundant fields, the change would have to be made in each row corresponding to the gene in question. If the change is not made in all relevant rows, an inconsistency arises in the data. In database circles, this inconsistency caused by unnecessary data duplication is called an update anomaly.
Another problem with spreadsheets is they are fundamentally single-user data sources. Only one user may enter data into a spreadsheet at a time. If multiple users must contribute data to a data source housed in a spreadsheet, a single curator must be designated. If multiple copies of a spreadsheet have been distributed, and each has been edited and added to by a different curator, it will be a substantial task to harmonize disagreements among the versions when a single canonical version is desired. The spreadsheet itself offers no help in this matter.
Finally, search methods over data stored in spreadsheets are limited to simple text searches over the entire spreadsheet; complex combinations of search conditions, such as “return serotonin receptors that have SNPs but do not have gene expression results” are not permitted. Additional limitations of text searches are presented in the next section.
Semi-Structured Text Files
Semi-structured text files, that is, text files containing a more or less regular series of labels and associated values, have data management limitations similar to spreadsheets. A prominent example is the GenBank sequence annotation flat files [10]. It should be noted that the National Center for Biotechnology Information (NCBI) does not store its data in flat file format; rather, the GenBank flat file format is simply a report format based on the structured ASN.1 data representation [11].
An advantage of the semi-structured text format is that it permits more complex, hierarchical (tree-like) structures to be represented. A sequence has multiple references, each of which has multiple authors. Text files are also perhaps the most portable of formats—anyone with a text editor program can view and edit them (unless the file size exceeds the limit of the editor’s capability). However, most text editors provide no data validation features. Like spreadsheets, they are not oriented toward use by multiple concurrent users and provide little help in merging or harmonizing multiple copies that have diverged from an original canonical version. Without writing an indexing program, searching a text file is very inefficient because the entire file must be read sequentially, looking for a match to the user’s input. Further, it is impossible to specify which part of the flat file entry is to be matched. If a user wants to find mammalian sequences, there is no way to limit the search to the section organism of the file to speed the search. As with spreadsheets, full-text searches over text files do not support complex combinations of search conditions. Full-text searches may also result in incorrect data retrieval. For example, consider a flat-file textual data source of human genes and their mouse orthologues, both of which have chromosomal locations. Suppose the user wants to “find all human genes related to mouse orthologues on mouse chromosome 10“; simple text-searching permits no way of specifying that the match to chromosome 10 should refer to the human gene and not the mouse gene. Finally, text editors provide no easy means of retrieving associated data from two related text data sources at once, for example, a GenBank entry and its associated Swiss-Prot entry. More sophisticated search capability over semi-structured, text-formatted data sources is provided by systems like LION Biosciences’ Sequence Retrieval System (SRS) [12] (presented in Chapter 5); however, such read-only indexing systems do not provide tools for data validation during curation or solve the multi-user concurrency problem, and they have limited power to compensate for data irregularities in the underlying text files.
3.2.3 Traditional Database Management
This discussion of the limitations of spreadsheets and flat files points toward the advantages of traditional data management approaches. The most mature of these, relational technology, was conceived in 1970 in a seminal paper by E. F. Codd [13]. In the succeeding 30 years, the technology has become very mature and robust, and a great deal of innovative thought has been put into making data retrieval faster and faster. For example, a great step forward was cost-based optimization, or planning a query based on minimizing the expense to execute it, invented in 1979 by Patricia Selinger [14]. Similarly, because relational technology was originally developed for business systems with a high volume of simultaneous inserts, updates and deletes, its ability to accommodate multiple concurrent users is highly advanced.
The Relational Model
A data model is the fundamental abstraction through which data is viewed. Although the terms are often confused, a data model is not the same as a schema, which represents the structure of a particular set of data. The basic element of the relational data model is a table (or relation) of rows (or tuples) and columns (or attributes). A representation of gene expression data in tabular fashion means the relational data model is being used. A particular relational schema might contain a gene table whose columns are GenBank accession number, Swiss-Prot accession number, description, chromosomal location, Pfam classification, CDS sequence, peptide sequence, GO annotation, gene expression results, SNPs, date_entered, and date_modified; and a gene expression table whose columns are GenBank_accession number, tissue_ID, and intensity_value.
A number of basic operations are defined on relations, expressed by the relational algebra operators [2]:
![]() Projection (Π) produces from a relation R a new relation (noted ΠR) that has only some of R’s columns. In the example, the projection operator might return only the GenBank and Swiss-Prot accession numbers of the genes in the table.
Projection (Π) produces from a relation R a new relation (noted ΠR) that has only some of R’s columns. In the example, the projection operator might return only the GenBank and Swiss-Prot accession numbers of the genes in the table.
![]() Selection (σ) produces from a relation R a new relation (noted σR) with a subset of R‘s rows. For example, this could be the genes that have a Pfam protein kinase domain.
Selection (σ) produces from a relation R a new relation (noted σR) with a subset of R‘s rows. For example, this could be the genes that have a Pfam protein kinase domain.
![]() The union (∪) of two relations R and S (noted R ∪ S) is the set of rows that are in R or S or both. (R and S must have identical sets of attributes.) For example, if there were 24 separate tables of genes, one for each human chromosome, the union operator could be used to yield a single table containing all the genes in the genome.
The union (∪) of two relations R and S (noted R ∪ S) is the set of rows that are in R or S or both. (R and S must have identical sets of attributes.) For example, if there were 24 separate tables of genes, one for each human chromosome, the union operator could be used to yield a single table containing all the genes in the genome.
![]() The difference operation noted R—S of two relations R and S is the set of elements that are in R but not in S; for example, this could be the set of GenBank accession numbers that appear in the genes table but are not present in the gene expression table.
The difference operation noted R—S of two relations R and S is the set of elements that are in R but not in S; for example, this could be the set of GenBank accession numbers that appear in the genes table but are not present in the gene expression table.
![]() The join (
The join (![]() ) of two relations R and S (noted R
) of two relations R and S (noted R ![]() S) is a relation consisting of all the columns of R and S, with rows from R and S paired if they agree on particular attribute(s) common to R and S, called the join attribute(s). For example, a user might join the genes table and the expression table on GenBank accession number, pairing genes with their expression results.
S) is a relation consisting of all the columns of R and S, with rows from R and S paired if they agree on particular attribute(s) common to R and S, called the join attribute(s). For example, a user might join the genes table and the expression table on GenBank accession number, pairing genes with their expression results.
The relational algebra operations are the building blocks that may be combined to form more complex expressions, or queries, that enable users to ask complex questions of scientific interest. For example, the following query involves projection, selection, union, and join: “Retrieve the GenBank accession numbers, peptide sequence, and tissue IDs” [projection] “for all genes on any chromosome” [union] “that have associated SNPs” [selection] “and show expression” [join] “in central nervous system tissue” [selection].
A key element of the relational approach is enabling users to describe the behavior they want to ensure or the results they want to retrieve, rather than requiring them to write a program that specifies, step by step, how to obtain the results or ensure the behavior. The Structured Query Language (SQL) [2], the language through which users pose questions to a relational database and specify constraints on relational data, is thus declarative rather than procedural. Through declarative statements, users can specify that a column value may not be null, that it must be unique in its table, that it must come from a predefined set or range of values, or that it must already be present in a corresponding column of another table. For example, when adding an expression result, the gene used must already be registered in the gene table. Through declarative queries, users can ask complex questions of the data involving many different columns in the database at once, and because relational tables may be indexed on multiple columns, such searches are fast. Advanced search capabilities permit defining subsets of the database and then counting or averaging numeric values over the subset. An example would be listing all tissues sampled and the average expression value in each over a set of housekeeping genes. Performing such computations over subsets of tables is called aggregation, and functions like count, average, minimum, and maximum are aggregate functions.
Finally, because it is easy to define multiple related tables in a relational database, a user may define separate tables for genes and their aliases, permitting fast searches over multiple aliases and eliminating the need for users to know what type of alias they are searching with, that is, where it comes from (Swiss-Prot, GenBank, etc.). There are two main disadvantages of relational databases when compared to flat file data sources and spreadsheets: Specialized software is required to query the data, and free text searches of the entire entry are not supported in a traditional relational database.
A criticism sometimes made of the relational data model is that it is not natural to model complex, hierarchically structured biological objects as flat, relational tables. For example, an annotated sequence, as represented in GenBank, is a rich structure. The systems in the BioKleisli family (see Chapter 6) address this issue by defining their basic operations on nested relations, that is, relations whose attributes can themselves be relations. Another approach to management of hierarchically structured data is to represent it in eXtensible Markup Language (XML) [15], a structured text data exchange format based on data values combined with tags that indicate the data’s structure. Special-purpose XML query languages are in development that will enable users to pose complex queries against XML databases and specify the desired structure of the resulting data [16].
Use Case: Transforming Database Structure
Often, transformation of database structures is necessary to enable effective querying and management of biological data. Many venerable data sources no longer represent biological objects optimally for the kinds of queries investigators typically want to pose. For example, it has often been noted that GenBank is sequence-centric, not gene-centric, so queries concerning the structure of individual genes are not easy to express. In contrast, Swiss-Prot is sequence-centric, not domain-centric, so it is rather awkward to ask for proteins with carbohydrate features in a certain domain because all these features are represented in terms of the sequence as a whole.
To illustrate one method of handling data transformations, consider a very simple gene table with attributes GenBank accession number, SwissProt accession number, and sequence. It might be advantageous to enable users to retrieve sequences by accession number without knowing where the accession number originated (GenBank or Swiss-Prot). Creating a separate table for aliases is one solution, particularly if each gene has many different accession numbers, including multiple accession numbers from the same original data source. Another way to permit this search is to transform the database into the following schema: accession number and sequence. This transformation can be accomplished by retrieving all the GenBank accession numbers and their associated sequences, then retrieving all the SwissProt accession numbers and their associated sequences, and finally doing a union of those two sets. The formula or expression that defines this transformed relation is called a view. This expression may be used to create a new table, called a materialized view, which exists separately from the original table, so that changes to the original are not applied to the new table. If the expression is not used to create a new table, but only to retrieve data from the original table and transform it on the fly, it is a non-materialized view, or simply a view.
Recall the critique that it is not natural to model complex, hierarchically structured biological objects as flat, relational tables. A user might choose relational database technology for storing and managing data due to its efficiency, maturity, and robustness but still wish to present a hierarchical view of the data to the user, one that more closely matches biological concepts. This (non-materialized) view may be accomplished by means of a conceptual schema layered on top of the relational database. The biological object layers of Transparent Access to Multiple Bioinformatics Information Sources (TAMBIS) (see Chapter 7) and the Acero Genome Knowledge Platform [17] are efforts in this direction.
3.3 DIMENSIONS DESCRIBING THE SPACE OF INTEGRATION SOLUTIONS
There is nearly universal agreement in the bioinformatics and genomics communities that scientific investigation requires an integrated view of all relevant data. A general discussion of the scope of biological data integration, as well as the obstacles that currently exist for integration efforts, is presented in Chapter 1 of this book. The typical bioinformatics practitioner encounters data in a wide variety of formats, as Chapter 2 presents, including relational databases, semi-structured flat files, and XML documents. In addition, the practicing bioinformatician must integrate the results of analytical applications performing such tasks as sequence comparison, domain identification, motif search, and phylogenetic classification. Finally, Internet sites are also critical due to the traditional importance of publicly funded, public domain data at academic and government Web sites, whether they are central resources or boutique data collections targeting specific research interests. These Internet resources often provide specialized search functionality as well as data, such as the Basic Local Alignment Search Tool (BLAST) at NCBI [18] and the Simple Modular Architecture Research Tool (SMART) at the European Molecular Biology Laboratory (EMBL) [19]. A bioinformatics integration strategy must make sure this specialized search capability is retained.
3.3.1 A Motivating Use Case for Integration
To motivate the need for an integration solution, consider the following use case: “Retrieve sequences for all human expressed sequence tags (ESTs) that by BLAST are >60% identical over >50 amino acids to mouse channel genes expressed in central nervous system (CNS) tissue.” For those less familiar with biological terms, a channel gene is a gene coding for a protein that is resident in the membrane of a cell and that controls the passage of ions (potassium, sodium, calcium) into and out of the cell. The channels open and close in response to appropriate signals and establish ion levels within the cell. This is particularly important for neural network cells. The data sources used in this query are: the Mouse Genome Database (MGD) at the Jackson Laboratory in Bar Harbor, Maine [20]; the Swiss-Prot protein sequence data source at the Swiss Institute for Bioinformatics, and the BLAST search tool and the GenBank nucleotide sequence data collection at NCBI. The data necessary to satisfy this query are split, or distributed, across multiple data sources at multiple sites. One way to integrate these data sources is to enable the user to access them as if they were all components of a single, large database with a single schema. This large global schema is an integrated view of all the local schemas of the individual data sources. Producing such a global schema is the task of schema integration.
This example illustrates six dimensions for categorizing integration solutions:
![]() Is data accessed via browsing or querying?
Is data accessed via browsing or querying?
![]() Is access provided via declarative or procedural code?
Is access provided via declarative or procedural code?
![]() Is the access code generic (used for all similar data sources) or hard-coded for the particular source?
Is the access code generic (used for all similar data sources) or hard-coded for the particular source?
![]() Is the focus on overcoming semantic heterogeneity (heterogeneity of meaning) or syntactic heterogeneity (heterogeneity of format)?
Is the focus on overcoming semantic heterogeneity (heterogeneity of meaning) or syntactic heterogeneity (heterogeneity of format)?
![]() Is integration accomplished via a data warehouse or a federated approach?
Is integration accomplished via a data warehouse or a federated approach?
![]() Is data represented in a relational or a non-relational data model?
Is data represented in a relational or a non-relational data model?
As will become evident, some approaches will be better suited to addressing this particular use case than others; this is not intended to prejudice but to clarify the differences among the approaches. The rest of Section 3.3 discusses various alternative approaches to addressing this motivating use case.
3.3.2 Browsing vs. Querying
The relationship between browsing and querying is similar to the relationship in library research between browsing the stacks and conducting an online search. Both are valid approaches with distinct advantages. Browsing, like freely wandering in the stacks, permits relatively undirected exploration. It involves a great deal of leg work, but it is the method of choice when investigators want to explore the domain of interest to help sharpen their focus. It is also well suited to retrieval of a single Web page by its identifier or a book by its call number. On the other hand, querying, like online searching, permits the formulation of a complex search request as a single statement, and its results are returned as a single collated set. Both browsing and querying allow the user to select a set of documents from a large collection and retrieve them. However, browsing stops at retrieval, requiring manual navigation through the resulting documents and related material via static hyperlinks. Querying goes further than retrieval: It accesses the content [21] of the resulting documents, extracts information and manipulates it, for example, dropping some items and performing computations on others. Querying thus makes very efficient use of human time and is the method of choice when an investigator’s interests are already focused, especially if aggregations over subsets of data are involved.
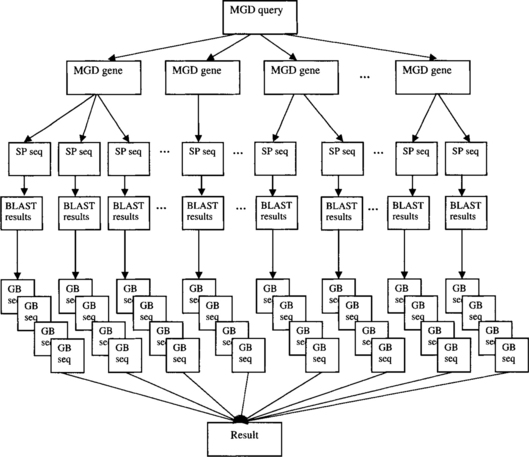
While the motivating use case may be successfully addressed using the browsing approach, it is tedious, error-prone, and very cumbersome, involving an average of 70 BLAST result sets consisting of up to 500 EST hits each. In the browsing approach, the user searches for channel sequences expressed in CNS tissues using the MGD query form. The result is 14 genes from 17 assays. The user then visits each gene’s MGD page. Assume that the user is only interested in Swiss-Prot sequences and that each gene has an average of five associated Swiss-Prot sequence entries. The user has to visit each sequence’s Swiss-Prot page, from which a BLAST search against gbest (the EST portion of GenBank) is launched. Each BLAST result must be inspected to eliminate non-human sequence hits and alignments that do not meet the inclusion criteria (>60% identity over >50 amino acids) and to eliminate duplicate ESTs hit by multiple Swiss-Prot sequences. Finally, the full EST sequences for all the hits that survive must be retrieved from GenBank. If the browsing approach was used to satisfy this query, these steps would then be repeated for each of the 14 genes returned by the initial query (Figure 3.1).
In a querying approach to this problem, a short SQL query is submitted to the query processor. The query processor visits MGD to identify channel genes expressed in CNS, and the Swiss-Prot Web site to retrieve their sequences. For each of these sequences, it launches a BLAST search against gbest, gathers the results, applies the stringency inclusion criteria, and finally retrieves the full-length EST sequences from GenBank (Figure 3.2).
3.3.3 Syntactic vs. Semantic Integration
As stated previously, syntactic integration addresses heterogeneity of form. GenBank is a structured file, MGD is a Sybase (relational) database, and BLAST is an analytical application. These differences in form are overcome in the browsing strategy by providing a Web-based front end to the sources and in the querying strategy by providing SQL access to all the sources. Contrariwise, semantic integration addresses heterogeneity of meaning. In GenBank, a gene is an annotation on a sequence, while in MGD a gene is a locus conferring phenotype (e.g., black hair, blindness). Neither of the integration approaches in this example specifically focuses on resolving this heterogeneity of meaning. They rely instead on the user’s knowledge of the underlying data sources to combine data from the sources in scientifically meaningful ways.
3.3.4 Warehouse vs. Federation
In a warehousing approach to integration, data is migrated from multiple sources into a single DBMS, typically a relational DBMS. As it is copied, the data may be cleansed or filtered, or its structure may be transformed to match the desired queries more closely. Because it is a copy of other data sources, a warehouse must be refreshed at specified times—hourly, daily, weekly, monthly, or quarterly. A data warehouse may contain multiple data marts, subset warehouses designed to support a specific activity or inquiry.
While a warehouse replicates data, a federated approach leaves data in its native format and accesses it by means of the native access methods. In the previous example, the querying approach is a federated approach—it accesses MGD as a Sybase database, Swiss-Prot and GenBank as Web sites, and BLAST via runtime searches, and it integrates their results using complex software known as middleware. An alternative demonstration of the querying approach could have imported GenBank, Swiss-Prot, MGD, and the results of BLAST searches into Sybase, Oracle, or IBM DB2 database systems and executed the retrievals and filtering there. This would have been an example of the warehousing approach.
3.3.5 Declarative vs. Procedural Access
As discussed previously, declarative access means stating what the user wants, while procedural access specifies how to get it. The typical distinction opposes the use of a query language (e.g., SQL) and writing access methods or sub-routines in Perl, Java, or other programming languages to access data. In the motivating use case in Section 3.3.1, the querying approach uses the SQL query language. Alternatively, Perl [22] sub-routines or object methods that extract data from MGD, Swiss-Prot, and GenBank and run the necessary BLAST searches could have performed the task.
3.3.6 Generic vs. Hard-Coded
The federated approach in the previous example was generic; it assumed the use of a general-purpose query execution engine and general purpose wrappers (software modules tailored to a particular family of data sources) for data access. An example of a hard-coded approach to the problem would be writing a special purpose Perl script to retrieve just the information needed to answer this particular question. A generic system enables users to ask numerous queries supporting a variety of scientific tasks, while a hard-coded approach typically answers a single query and supports users in a single task. Generic approaches generally involve higher up-front development costs, but they can pay for themselves many times over in flexibility and ease of maintenance because they obviate the need for extensive programming every time a new research question arises.
3.3.7 Relational vs. Non-Relational Data Model
Recall that a data model is not a specific database schema, but rather something more abstract: the way in which data are conceptualized. For example, in the relational data model, the data are conceptualized as a set of tables with rows and columns. Oracle, Sybase, DB2, and MySQL are all DBMSs built on the relational model. In data management systems adhering to a non-relational data model, data may be conceptualized in many different ways, including hierarchical (tree-like) structures, ASCII text files, or Java or Common Object Request Broker Architecture (CORBA) [23] objects. In the motivating example, MGD is relational, and the other sources are non-relational.
3.4 USE CASES OF INTEGRATION SOLUTIONS
The motivating use case in Section 3.3.1 permitted a brief outline of the six dimensions for categorizing integration solutions. To further elucidate these dimensions and demonstrate their use, this section describes each dimension in greater detail, presents a prototypical featured solution, and categorizes the featured integration solution on all six dimensions.
3.4.1 Browsing-Driven Solutions
As in the previous example, in a browsing approach users are provided with interactive access to data, allowing them to step sequentially through the exploratory process. A typical browsing session begins with a query form that supports a set of pre-defined, commonly posed queries. After the user has specified the parameters of interest and the query is executed, a summary screen is typically returned. From here the user may drill down, one by one, into the individual objects meeting the search criteria and from there view related objects by following embedded links, such as hypertext markup language (HTML) or XML hyperlinks. The data source(s) underlying a browsing application may be warehoused or federated, and relational or non-relational. Browsing applications are ubiquitous on the Internet; examples are Swiss-Prot and the other data collections on the Expert Protein Analysis System (ExPASy) server at the Swiss Institute of Bioinformatics [24], the FlyBase Web site for Drosophila genetics [25, 26], and the featured example, the Entrez Web site at NCBI [10].
Browsing Featured Example: NCBI Entrez
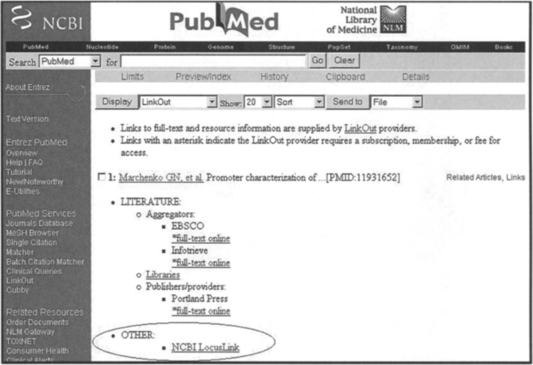
As an example of the browsing approach, consider the following query: “Find in PubMed articles published in 2002 that are about human metalloprotease genes and retrieve their associated GenBank accession numbers and sequences.” The sequence of steps in answering this query is shown in Figures 3.3 through 3.7. First the user enters the Entrez Boolean search term “metalloprotease AND human AND 2002 [pdat]” in the PubMed online query form [7]. The result is a summary of qualifying hits; there were 1054 in December 2002 (Figure 3.3). From here, the user can visit individual PubMed entries (Figure 3.4), read their abstracts, check for a GenBank sequence identifier in the secondary source ID attribute (Figure 3.5), and visit the associated GenBank entry to retrieve the sequence.
Alternatively, the user can take advantage of the LinkOut option on the PubMed entry page (Figures 3.4 and 3.6), which enables access to the sequence information provided by LocusLink (Figure 3.7). Notice that there are many more navigation paths to follow via hyperlinks than are described here; a strength of the browsing approach is that it supports many different navigation paths through the data.
The categorization of Entrez based on the six dimensions is given in Table 3.1 on page 56.
TABLE 3.1
Entrez categorization with respect to the six dimensions of integration.
| Browsing | Querying |
| Interactive Web browser access to data | No querying capability |
| Semantic | Syntactic |
| No semantic integration | Provides access to nucleotide and protein sequence, annotation, MEDLINE abstracts, etc. |
| Warehouse | Federation |
| Provides access to data sources at NCBI | No federation |
| Declarative Access | Procedural Access |
| No declarative access | Access via Entrez Programming Utilities (E-utilities) |
| Generic | Hard-Coded |
| Not generic | Hard-coded for NCBI sources only Links are hard-coded indices |
| Relational Data Model | Non-Relational Data Model |
| Relational data model not used | Data stored in the ASN.1 complex-relational data model |
3.4.2 Data Warehousing Solutions
In the data warehousing approach, data is integrated by means of replication and storage in a central repository. Often data is cleaned and/or transformed during the loading process. While a variety of data models are used for data warehouses, including XML and ASN.1, the relational data model is the most popular choice (e.g., Oracle, Sybase, DB2, MySQL). Examples of the integration solutions following the data warehousing approach include Gene Logic’s GeneExpress Database (presented in Chapter 10) [27], the Genome Information Management System of the University of Manchester [28], the data source underlying the GeneCards Web site at the Weizmann Institute in Israel [29, 30], and AllGenes [31], which will serve as the featured example.
Warehousing Featured Example: AllGenes
A research project of the Computational Biology and Informatics Laboratory at the University of Pennsylvania, AllGenes is designed to provide access to a database integrating every known and predicted human and mouse gene, using only publicly available data. Predicted human and mouse genes are drawn from transcripts predicted by clustering and assembling EST and messenger RNA (mRNA) sequences. The focus is on integrating the various types of data (e.g., EST sequences, genomic sequence, expression data, functional annotation). Integration is performed in a structured manner using a relational database and controlled vocabularies and ontologies [32]. In addition to clustering and assembly, significant cleansing and transformation are done before data is loaded onto AllGenes, making data warehousing an excellent choice.
A sample query for AllGenes is the following: “Show me the DNA repair genes that are known to be expressed in central nervous system tissue.” The query is specified and run using a flexible query-builder interface (Figure 3.8), yielding a summary of qualifying assemblies (Figure 3.9). From the query result page the user can visit a summary page for each qualifying assembly, which includes such valuable information as predicted GO functions; hyperlinks to GeneCards, the Mouse Genome Database (MGD), GenBank, ProDom, and so on; Radiation Hybrid (RH) Map locations; the 10 best hits against the GenBank non-redundant protein database (nr); and the 10 best protein domain/motif hits.
The categorization of AllGenes based on the six dimensions is given in Table 3.2 on page 58.
TABLE 3.2
AllGenes categorization with respect to the six dimensions of integration.
| Browsing | Querying |
| Interactive Web browser access to data | Limited querying capability via parameterized query builder |
| Semantic | Syntactic |
| Ontologies for semantic integration | Data warehousing for syntactic integration |
| Warehouse | Federation |
| Data stored in relational warehouse | Not a federation |
| Declarative Access | Procedural Access |
| Under the covers; users use parameterized query builder | No procedural access |
| Generic | Hard-Coded |
| Information not available | Information not available |
| Relational Data Model | Non-Relational Data Model |
| Data stored in Oracle DBMS | Not used |
3.4.3 Federated Database Systems Approach
Recall that in a federated approach, data sources are not migrated from their native source formats, nor are they replicated to a central data warehouse. The data sources remain autonomous, data is integrated on the fly to support specific queries or applications, and access is typically through a declarative query language. Examples of federated systems and their data models include complex-relational systems, such as BioKleisli/K2 (Chapter 8) and its cousin GeneticXchange’s K1 (see Chapter 6), object-relational systems (OPM/TINet) [33], and IBM’s relational system Discovery Link, which is detailed in Chapter 10 and will serve as the featured example [34].
Federated Featured Example: DiscoveryLink
The motivating use case of Section 3.3.1 is a good fit for a federated approach like DiscoveryLink’s. DiscoveryLink provides transparency: The federation of diverse types of data from heterogeneous sources appears to the user or the application as a single large database, in this case a relational database. The SQL query language is supported over all the federated sources, even if the underlying sources’ native search capabilities are less full-featured than SQL; a single federated query, as in the earlier motivating example, typically combines data from multiple sources. Similarly, specialized non-SQL search capabilities of the underlying sources are also available as DiscoveryLink functions.
The architecture of DiscoveryLink appears in Chapter 10 (Figure 10.1). At the far right are the data sources. To these sources, DiscoveryLink looks like an application—they are not changed or modified in any way. DiscoveryLink talks to the sources using wrappers, which use the data source’s own client-server mechanism to interact with the sources in their native dialect. DiscoveryLink has a local catalog in which it stores information (meta-data) about the data accessible (both local data, if any, and data at the back end data sources). Applications of DiscoveryLink manipulate data using any supported SQL Application Programming Interface (API); for example, Open Database Connectivity (ODBC) or Java DataBase Connectivity (JDBC) are supported, as well as embedded SQL. Thus a DiscoveryLink application looks like any normal database application.
The categorization of DiscoveryLink based on the six dimensions is given in Table 3.3.
3.4.4 Semantic Data Integration
Recall that semantic data integration focuses on resolving heterogeneity of meaning, while syntactic data integration focuses on heterogeneity of form. In a volume on management of heterogeneous database systems, Kashyap and Sheth write:
In any approach to interoperability of database systems [database integration], the fundamental question is that of identifying objects in different databases that are semantically related and then resolving the schematic [schema-related] differences among semantically related objects. [35]
This is the fundamental problem of semantic data integration. The same protein sequence is known by different names or accession numbers (synonyms) in GenBank and Swiss-Prot. The same mouse gene may be represented as a genetic map locus in MGD, the aggregation of multiple individual exon entries in GenBank, and a set of EST sequences in UniGene; in addition, its protein product may be an entry in Swiss-Prot and its human orthologues may be represented as a disease-associated locus in Online Mendelian Inheritance in Man (OMIM) [36]. Semantic integration also deals with how different data sources are to be linked together. For example, according to documentation at the Jackson Lab Web site [37], MGD links to Swiss-Prot through its marker concept, to RatMap [38] through orthologues, to PubMed through references, and to GenBank through either markers (for genes) or molecular probes and segments (for anonymous DNA segments). Finally, a schema element with the same names in two different data sources can have different semantics and therefore different data values. For example, retrieving orthologues to the human BRCA1 gene in model organisms from several commonly used Web sites yields varying results: GeneCards returns the BRCA1 gene in mouse and C. elegans; MGD returns the mouse, rat, and dog genes; the Genome DataBase (GDB) [39, 40] returns the mouse and drosophila genes; and LocusLink returns only the mouse gene.
Approaches to semantic integration in the database community generally center on schema integration: understanding, classifying, and representing schema differences between two disparate databases. For example, in capturing the semantics of the relationships between objects in multiple databases, Kashyap and Sheth describe work on understanding the context of the comparison, the abstraction relating the domains of the two objects, and the uncertainty in the relationship [35].
Bioinformatics efforts at semantic integration have largely followed the approach of the artificial intelligence community. Examples of such semantic integration efforts are the Encyclopedia of Escherichia coli genes and metabolism (EcoCyc) [41], GO, and TAMBIS, the featured example and the subject of Chapter 7 of this book.
Semantic Integration Featured Example: TAMBIS
The TAMBIS system is the result of a research collaboration between the departments of computer science and biological sciences at the University of Manchester in England. Its chief components are an ontology of biological and bioinformatics terms managed by a terminology server and a wrapper service that, as in DiscoveryLink, handles access to external data sources. An ontology is a rigorous formal specification of the conceptualization of a domain. The TAMBIS ontology (TaO) [42] describes the biologist’s knowledge in a manner independent of individual data sources, links concepts to their real equivalents in the data sources, mediates between (near) equivalent concepts in the sources, and guides the user to form appropriate biological queries. The TaO contains approximately 1800 asserted biological concepts and their relationships and is capable of inferring many more. Coverage currently includes proteins and nucleic acids, protein structure and structural classification, biological processes and functions, and taxonomic classification.
The categorization of TAMBIS based on the six dimensions is given in Table 3.4.
TABLE 3.4
TAMBIS categorization with respect to the six dimensions of integration.
| Browsing | Querying |
| Interactive browser | Limited querying capability via parameterized query builder |
| Semantic | Syntactic |
| According to TAMBIS’ authors, its | Integrates via its wrapper service |
| “big win” lies in the ontology | |
| Warehouse | Federation |
| Not used | Uses BioKleisli for federated integration |
| Declarative Access | Procedural Access |
| Uses the CPL query language, but users see | No procedural access |
| only the parameterized query builder | |
| Generic | Hard-Coded |
| Information not available | Information not available |
| Relational Data Model | Non-Relational Data Model |
| Relational data model not used | Object/complex-relational data model |
3.5 STRENGTHS AND WEAKNESSES OF THE VARIOUS APPROACHES TO INTEGRATION
This chapter has described multiple approaches to database integration in the bioinformatics domain and provided examples of each. Each of these approaches has strengths and weaknesses and is best suited to a particular set of integration needs.
3.5.1 Browsing and Querying: Strengths and Weaknesses
The strengths of a browsing approach are many. As noted previously, its interactive nature makes it especially well suited to exploring the data landscape when an investigator has not yet formulated a specific question. It is also well suited to retrieval of information about single objects and for optionally drilling down to greater levels of detail or for following hyperlinks to related objects. The ubiquity of the Internet makes Web browsers familiar to even the most inexperienced user.
The weaknesses of a browsing approach are the flip side of its strengths. Because it is fundamentally based on visiting single pages containing data on a single object, it is not well suited to handling large data sets or to performing a large, multi-step workflow including significant processing of interim results. Its flexibility is also limited, as the user is confined to the query forms and navigation paths the application provides.
The strengths of a querying approach are the natural opposite of those of the browsing approach. Because it is based on specifying attributes of result sets via a query language, often with quite complex search conditions, the querying approach is well suited to multi-step workflows resulting in large result sets. This approach is also flexible, allowing the user to specify precisely inclusion and exclusion criteria and noting which attributes to include in the final result set. Contrariwise, the querying approach is not as well suited to the exploration or manual inspection of interim results, and the need to specify desired results using query language syntax requires more computational sophistication than many potential users possess.
3.5.2 Warehousing and Federation: Strengths and Weaknesses
A major strength of a data warehousing approach is that it permits cleansing and filtering of data because an independent copy of the data is being maintained. If the original data source is not structured optimally to support the most commonly desired queries, a warehousing approach may transform the data to a more amenable structure. Copying remote data to a local warehouse can yield excellent query performance on the warehouse, all other things being equal. Warehousing exerts a load on the remote sources only at data refresh times, and changes in the remote sources do not directly affect the warehouse’s availability.
The primary weakness of the data warehousing approach is the heavy maintenance burden incurred by maintaining a cleansed, filtered, transformed copy of remote data sources. The warehouse must be refreshed frequently to ensure users’ access to up-to-date data; the warehousing approach is probably not the method of choice for integrating large data sources that change on a daily basis. Adding a data source to a warehouse requires significant development, loading, and maintenance overhead; therefore this approach is unlikely to scale well beyond a handful of data sources. Warehousing data may lose the specialized search capability of the native data sources; an example would be specialized text searching over documents or sub-structure searching over chemical compound data collections.
A major strength of the federated approach is that the user always enjoys access to the most up-to-date data possible. While connectivity to remote sources requires some maintenance, the burden of adding and maintaining a new data source is considerably less than in the warehousing case. The federated approach scales well, even to very large numbers of data sources, and it readily permits new sources to be added to the system on a prototype or trial basis to evaluate their potential utility to users. In a fast-paced, ever-changing field like bioinformatics, this nimbleness is invaluable. The federated approach meshes well with a landscape of many individual, autonomous data sources, which the bioinformatics community currently boasts. Finally, a federated system can provide access to data that cannot be easily copied into a warehouse, such as data only available via a Web site.
Any data cleansing must be done on the fly, for a federation accesses remote data sources in their native form. The members of the federation must be able to handle the increased load put on them by federated queries, and if network bandwidth is insufficient, performance will suffer.
3.5.3 Procedural Code and Declarative Query Language: Strengths and Weaknesses
Procedural code may be tuned very precisely for a specific task. There are virtually no limitations on its expressive power; however, this very strength can make it difficult to optimize. Ad hoc inquiries can be difficult to support, and extending the system to handle additional sources or additional queries can be difficult.
Declarative languages are flexible and permit virtually unlimited ad hoc querying. Queries expressed in a declarative language are relatively easy to program and maintain due to their small size and economy of expression. Sometimes, however, their simplicity is misleading; for example, it is easy to write a syntactically correct SQL query, but the results returned may not be what was intended because the query was written using the wrong constructs for the desired meaning. Finally, some programming tasks are much more easily written in a procedural language than a declarative one; the classic example is recursive processing over tree-like structures.
3.5.4 Generic and Hard-Coded Approaches: Strengths and Weaknesses
Generic coding is generally acknowledged to be desirable, where practicable, due to its extensibility and maintainability and because it facilitates code re-use. It does, however, yield a greater up-front cost than programming hard-coded for a specific task, and sometimes schedules do not permit this up-front expenditure. If the instances being generalized are not sufficiently similar, the complexity of generic code can be prohibitive.
Hard-coding permits an application to be finely tuned to optimize for a specific critical case, potentially yielding very fast response times; this approach may be the preferred strategy when only a limited set of queries involving large datasets is required. In the absence of an already existing generic system, it is generally quicker to prototype rapidly by hard coding. On the other hand, code with many system-specific assumptions or references can be difficult to maintain and extend. Adding a new data source or even a new query often means starting from scratch.
3.5.5 Relational and Non-Relational Data Models: Strengths and Weaknesses
The relational data model is based on a well-understood, theoretically rock-solid foundation. Relational technology has been maturing for the past 30 years and can provide truly industrial-strength robustness and constant availability. Relational databases prevent anomalies while multiple users are reading and writing concurrently, thus safeguarding data integrity. Optimization of queries over relational databases has been developed and honed for decades. The SQL query language is powerful and widely used, so SQL programmers are relatively easy to find. However, the relational model is based on tables of rows and columns, and several individual tables are typically required to represent a single complex biological object.
Hierarchical non-relational data models seem to be a more natural fit for complex scientific objects. However, this technology is still quite immature, and standard database desiderata such as cost-based query optimization, data integrity, and multi-user concurrency have been hard to attain because of the increased complexity of the non-relational systems.
3.5.6 Conclusion: A Hybrid Approach to Integration Is Ideal
Considering the variety of integration needs in a typical organization, a hybrid approach to database integration is generally the best strategy. For data that it is critical to clean, transform, or hand curate, and for which only the best query performance is adequate, data warehousing is probably the best approach. If the warehouse is derived from data outside the organization, it is best if the original data source changes infrequently, so the maintenance burden in merging updates is not too onerous. Otherwise, the federated model is an excellent choice because of its relatively low maintenance cost and its extensibility and scalability. Federations allow easy prototyping and swapping of new data sources for old in evaluation mode, and they permit integration of external data that is not accessible for duplicating internally, such as data only available via Web sites. They also permit the integration of special purpose search algorithms such as sequence comparison, secondary structure prediction, text mining, clustering, chemical structure searching, and so forth. Wherever possible, strategies should be generic, except for one-time, one-use programs or where hard-coding is needed to fine tune a limited set of operations over a limited set of data.
Both browsing and querying interfaces are important for different levels of users and different needs. For access to data in batch mode, the most common queries can be pre-written and parameterized and offered to users via a Web form-based interface. Both semantic and syntactic data integration are needed, although semantic integration is just beginning to be explored and understood.
Due to the maturity of the technology and its industrial strength, the relational data model is currently the method of choice for large integration efforts, both warehousing and federation. A middle software layer may be provided to expose biological objects to users, as mentioned previously. But based on the current state of the industry, the underlying data curation, storage, querying planning, and optimization are arguably best done in relational databases.
3.6 TOUGH PROBLEMS IN BIOINFORMATICS INTEGRATION
In spite of the variety of techniques and approaches to data integration in bioinformatics, many tough integration problems remain. These include query processing in a federated system when some members of the federation are inaccessible; universally accepted standards of representation for central biological concepts such as gene, protein, transcript, sequence, polymorphism, and pathway; and representing and querying protein and DNA interaction networks. This section discusses two additional examples of tough problems in bioinformatics integration: semantic query planning and schema management.
3.6.1 Semantic Query Planning Over Web Data Sources
While the TAMBIS and GO projects have made an excellent start in tackling the semantic integration problem, more remains to be done. TAMBIS and GO have focused on building ontologies and controlled vocabularies for biological concepts. Another fruitful area of investigation in semantic integration is using knowledge of the semantics of data sources to generate a variety of alternative methods of answering a question of scientific interest, thus freeing the user from the need to understand every data source in detail [43].
Recall that in accessing multiple data sources there are usually multiple ways of executing a single query, or multiple query execution plans. Each may have a different execution cost, as discussed in Section 3.2.1. Similarly, there may be multiple data sources that can be used to arrive at an answer to the same general question, though the semantics of the result may differ slightly. A semantic query planner considers not only the cost of different execution plans but also their semantics and generates alternate paths through the network of interconnected data sources. The goal is to help the user obtain the best possible answers to questions of scientific interest.
Web sources are ubiquitous in bioinformatics, and they are connected to each other in a complex tangle of relationships. Links between sources can be either explicit hypertext links or constructed calls in which an identifier for a remote data source may be extracted from a Web document and used to construct a Uniform Resource Identifier (URI) to access the remote source.
Not all inter-data source links are semantically equivalent. For example, there are two ways of navigating from PubMed to GenBank: through explicit occurrences of GenBank accession numbers within the secondary source identifier (SI) attribute of the MEDLINE formatted entry and through the Entrez Nucleotide Link display option. Following these two navigation paths does not always produce the same set of GenBank entries: For example, for the PubMed entry with ID 8552191, there are four embedded GenBank accession numbers, while the Nucleotide Links option yields 10 sequence entries (the four embedded entries plus related RefSeq entries) [43].
To generate alternate plans, a semantic query planner requires knowledge of certain characteristics of Web sources, including their query and search capabilities, the links between sources, and overlaps between the contents of data sources. An example of modeling a subset of the search capabilities of PubMed is as follows:
1. Search by key (PubMedID or MedlineID), returning a single entry. Single or multiple bindings for PubMedID or MedlineOD are accepted.
2. Search by phrase, returning multiple entries. For example, the search term gene expression performs an untyped text search; Science [journal] returns all articles from Science; and 2001/06:2002 [pdat] returns articles published since June 2002.
To further illustrate semantic query planning, consider the following query: “Given a Human Genome Organization (HUGO) name, retrieve all associated PubMed citations.” There are at least three plans for this query:
1. Search PubMed directly for the HUGO name using the second search capability above.
2. Find the GeneCards entry for the HUGO name and follow its link to PubMed publications.
3. From the GeneCards entry for the HUGO name, follow the links to the Entrez RefSeq entry and extract the relevant PubMed identifiers.
These three plans all return different answers. For example, given the HUGO name BIRC1 (neuronal apoptosis inhibitory protein), plan 1 returns no answer, plan 2 returns two answers (PubMed identifiers 7813013 and 9503025), and plan 3 returns five answers (including the two entries returned by plan 2) [43].
In summary, a query planner who took advantage of semantic knowledge of Web data sources and their search capabilities would first identify that there are multiple alternate sources and capabilities to answer a query. Then, semantic knowledge would be used to determine if the results of each alternate plan would be identical. Finally, such a planner might suggest these alternate plans to a user, whose expert judgment would determine which plan was the most suitable to the scientific task. The user would be freed to focus on science instead of on navigating the often treacherous waters of data source space. Semantic query planning will be addressed in more detail in Section 4.4.2 in Chapter 4.
3.6.2 Schema Management
A schema management system supports databases and information systems as they deal with a multitude of schemas in different versions, structure, semantics, and format. Schema management is required whenever data is transformed from one structure to another, such as publishing relational data as XML on a Web site, restructuring relational data in hierarchical form for a biological object conceptual view layer, and integrating overlapping data sets with different structures, as needed in a merger of two large pharmaceutical companies. The system developed by the Clio research project [44] is an example of a basic schema management system with plans for development in the direction to be described.
The six building blocks of a schema management system are listed below. They will be defined and illustrated through three use cases.
![]() Schema association/schema extraction
Schema association/schema extraction
![]() Schema versioning/schema evolution
Schema versioning/schema evolution
![]() Schema mapping/query decomposition
Schema mapping/query decomposition
Use Case: Data Warehousing
As described earlier, data warehousing is often used as an integration approach when the data must be extensively cleaned, transformed, or hand-curated. A warehouse may be built from a variety of data sources in different native formats. Schema association determines if these heterogeneous documents match a schema already stored in the schema manager. If no existing schema is found, schema extraction determines a new schema based on the data, for example, an XML document, and adds it to the schema manager. Schema integration helps develop a warehouse global schema accommodating all relevant data sources. As the warehouse evolves, schema mapping determines how to map between the schemas of newly discovered data sources and the warehouse’s global schema. Finally, data transformation discovers and executes the complex operations needed to clean and transform the source data into the global warehouse schema. The transformations generated would be specified in the XML query language (XQuery) or Extensible Stylesheet Language Transformations (XSLT) for XML data, and SQL for relational data.
Use Case: Query and Combine Old and New Data
Because bioinformatics is a young, research-oriented field, database schemas to hold lab notebook data change frequently as new experimental techniques are developed. Industry standards are still emerging, and they evolve and change rapidly. Suppose two Web sites at a large pharmaceutical company were using two different versions of the same database schema, but scientists wanted to query the old-version and new-version data sources in uniform fashion, without worrying about the schema versions. Schema evolution keeps track of schema versions and their differences. Query decomposition allows the user to query old and new documents in a single query, as if they all conformed to the latest schema version, using knowledge of the differences between versions.
Use Case: Data Federation
Assume a federated database system integrates relational (e.g., MGD, GO, GeneLynx [45]) and XML data sources (e.g., PubMed and the output of bioinformatics algorithms such as BLAST) and provides integrated SQL access to them. View building allows users to build customized views on top of relational and XML schemas using a graphical interface. Schema mapping provides knowledge about correspondences among the different sources. To respond efficiently to queries against these sources, view composition and query decomposition must use the correspondences gained through schema mapping and view building to issue the right queries to the right sub-systems. Finally, knowledge about the data sources’ capabilities and global query optimization allow the processor to push expensive operations to local sources as appropriate.
3.7 SUMMARY
Effective data management and integration are critical to the success of bioinformatics, and this chapter has introduced key concepts in these technical areas. While the wide and varied landscape of integration approaches can seem overwhelming to the beginner, this chapter has offered six dimensions by which to characterize current and new integration efforts: browsing/querying, declarative/procedural code, generic/hard-coded code, semantic/syntactic integration, data warehousing/federation, and relational/non-relational data model. Basic definitions and the relative strengths and weaknesses of a variety of approaches were explored through a series of use cases, which are summarized in Table 3.5. The optimal strategy for a given organization or research project will vary with its individual needs and constraints, but it will likely be a hybrid strategy, based on a careful consideration of the relative strengths and weaknesses of the various approaches. While many areas of data integration are solved or nearly so, tough, largely unsolved problems still remain. The chapter concluded by highlighting two of them: semantic query planning and schema management.
ACKNOWLEDGMENTS
Warm thanks are offered to my colleagues at IBM for many stimulating discussions and collegial support: Laura Haas, Peter Schwarz, Julia Rice, and Felix Naumann. Thanks also to Carole Goble and Robert Stevens of the University of Manchester for kindly providing materials on the TAMBIS project; Howard Ho and the IBM Clio team for their generous contributions to the schema management section; and Bill Swope for help on data warehousing. Finally, I thank the editors and reviewers for their patience and helpful suggestions.
REFERENCES
[1] Rashidi, H.H., Buehler, L.K. Bioinformatics Basics: Applications in Biological Science and Medicine. Boca Raton, FL: CRC Press; 2000.
[2] Ullman, J.D., Widom, J. A First Course in Database Systems. Upper Saddle River, NJ: Prentice Hall; 1997.
[3] Benson, D., Karsch-Mizrachi, I., Lipman, D., et al, GenBank. Nucleic Acids Research. 2003;31(no. 1):23–27. http://www.ncbi.nlm.nih.gov/Genbank
[4] Boeckmann, B., Bairoch, A., Apweiler, R., et al. The SWISS-PROT Protein Knowledgebase and Its Supplement TrEMBL in 2003. Nucleic Acids Research. 2003;31(no. 1):365–370.
[5] Bateman, A., Birney, E., Cerruti, L., et al. The Pfam Protein Families Database. Nucleic Acids Research. 2002;30(no. 1):276–280.
[6] Ashburner, M., Ball, C.A., Blake, J.A., et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nature Genetics. 2000;25(no. 1):25–29.
[7] Wheeler, D.L., Church, D.M., Lash, A.E., et al. Database Resources of the National Center for Biotechnology Information: 2002 Update. Nucleic Acids Research. 2002;30(no. 1):13–16.
[8] Kanehisa, M., Goto, S., Kawashima, S., et al. The KEGG Databases at GenomeNet. Nucleic Acids Research. 2002;30(no. 1):42–46.
[9] Microsoft Corporation. Microsoft Excel 2000, 1999.
[10] National Center for Biotechnology Information. The Entrez Search and Retrieval System. www.ncbi.nlm.nih.gov/Entrez. 2002.
[11] Ostell, J.M., Wheelan, S.J., Kans, J.A. The NCBI Data Model. Methods of Biochemical Analysis. 2001;43:19–43.
[12] Ezold, T., Argos, P. SRS: An Indexing and Retrieval Tool for Flat File Data Libraries. Computer Applications in the Biosciences. 1993;9(no. 1):49–57.
[13] Codd, E.F. A Relational Model of Data for Large Shared Data Banks. Communications of the ACM. 1970;13(no. 6):377–387.
[14] Selinger, P.G., Astrahan, M.M., Chamberlin, D.D., et al, Access Path Selection in a Relational Database Management System, Proceedings of the 1979 ACM SIGMOD International Conference on Management of Data. Boston, MA, May 30-June 1. ACM. 1979:23–34.
[15] October 6 Bray, T., Paoli, J., Sperberg-McQueen, C.M., et al, Extensible Markup Language (XML): World Wide Web Consortium (W3C) Recommendation. 2nd edition. 2000. http://www.w3.org/TR/REC/xml/html
[16] http://www.w3c.org/xme/query.
[17] The Acero Genome Knowledge Platform, http://www.acero.com.
[18] Altschul, S.F., Gish, W., Miller, W., et al. Basic Local Alignment Search Tool. Journal of Molecular Biology. 1990;215(no. 3):403–410.
[19] Letunic, I., Goodstadt, L., Dickens, N.J., et al. Recent Improvements to the SMART Domain-Based Sequence Annotation Resource. Nucleic Acids Research. 2002;30(no.l):242–244.
[20] Blake, J.A., Richardson, J.E., Bult, C.J., et al. The Mouse Genome Database (MGD): The Model Organism Database for the Laboratory Mouse. Nucleic Acids Research. 2002;30(no. 1):113–115.
[21] Lacroix, Z., Sahuget, A., Chandrasekar, R., Information Extraction and Database Techniques: A User-Oriented Approach to Querying the Web, Proceedings of the 1998 10th International Conference on Advanced Information Systems Engineering (CAiSE ’98). Pisa, Italy, June 8–12. 1998:289–304.
[22] Wall, L., Christiansen, T., Schwartz, R. Programming Perl, 2nd edition. Sebastopol, CA: O’Reilly and Associates; 1996.
[23] Mowbray, T.J., Zahavi, R. The Essential CORBA: Systems Integration Using Distributed Objects. New York: Wiley; 1995.
[24] The Expert Protein Analysis System (ExPASy) server at the Swiss Institute of Bioinformatics, http://www.expasy.ch.
[25] The FlyBase Web site for Drosophila genetics, http://flybase.bio.indiana.edu.
[26] Gelbart, W.M., Crosby, M., Matthews, B., et al. FlyBase: A Drosophilia Database. The FlyBase Consortium. Nucleic Acids Research. 1997;25(no. 1):63–66.
[27] Gene Logic’s GeneExpress Database, http://www.genelogic.com/genexpress.cfm.
[28] Cornell, M., Paton, N.W., Wu, S., et al. GIMS—A Data Warehouse for Storage and Analysis of Genome Sequence and Functional Data. In: Proceedings of the 2nd IEEE International Symposium on Bioinformatics and Bioengineering (BIBE). Rockville, MD: IEEE Press; 2001:15–22.
[29] GeneCards Web site at the Weizmann Institute in Israel, http://bioinfo.weizmann.ac.il/cards.
[30] Rebhan, M., Chalifa-Caspi, V., Prilusky, J., et al. GeneCards: A Novel Functional Genomics Compendium with Automated Data Mining and Query Reformulation Support. Bioinformatics. 1998;14(no. 8):656–664.
[31] version 5.0 The Computational Biology and Informatics Library, AllGenes: A Web Site Providing Access to an Integrated Database of Known and Predicted Human and Mouse Genes. Center for Bioinformatics, Unisversity of Pennsylvania; 2002. http://www.allgenes.org
[32] Davidson, S.B., Crabtree, J., Brunk, B.P., et al. K2/Kleisli and GUS: Experiments in Integrated Access to Genomic Data Sources. IBM Systems Journal. 2001;40(no. 2):512–531.
[33] Eckman, B.A., Kosky, A.S., Laroco, L.A., Jr. Extending Traditional Query-Based Integration Approaches for Functional Characterization of Post-Genomic Data. Bioinformatics. 2001;17(no. 7):587–601.
[34] Haas, L.M., Schwartz, P.M., Kodali, P., et al. DiscoveryLink: A System for Integrating Life Sciences Data. IBM Systems Journal. 2001;40(no. 2):489–511.
[35] Kashyap, V., Sheth, A. Semantic Similarities Between Objects in Multiple Databases. In: Elmagarmid A., Rusinkiewicz M., Sheth A., eds. Management of Heterogeneous and Autonomous Database Systems. 3rd edition. San Francisco: Morgan Kaufmann; 1999:57–89.
[36] Hamoush, A., Scott, A.F., Amberger, J., et al. Online Mendelian Inheritance in Man (OMIM), A Knowledge Base of Human Genes and Genetic Disorders. Nucleic Acids Research. 2002;30(no. 1):52–55.
[37] The Jackson Lab Web site, http://www.informatics.jax.org/mgihome/overview.shtml.
[38] RatMap, http://ratmap.gen.gu.se/.
[39] Genome DataBase (GDB), http://www.gdb.org.
[40] 1.13.1-1.13.12 Talbot, C.C., Jr., Cuticchia, A.J. Human Mapping Databases. In: Current Protocols in Human Genetics. New York: Wiley; 1999.
[41] Pellegrini-Toole, A., Bonavides, C., Gama-Castro, S. The EcoCyc Database. Nucleic Acids Research. 2002;30(no. 1):56–58.
[42] Baker, P.G., Gobel, C.A., Bechhofer, S., et al. An Ontology for Bioinformatics Applications. Bioinformatics. 1999;15(no. 6):510–520.
[43] Eckman, B.A., Lacroix, Z., Raschid, L. Optimized, Seamless Integration of Biomolecular Data. In: In proceedings of the 2nd IEEE International Symposium on Bioinformatics and Bioengineering (BIBE). Rockville, MD: IEEE; 2001:23–32.
[44] Miller, R.J., Haas, L.M., Yan, L., et al. The Clio Project: Managing Heterogeneity. ACM SIGMOD Record. 2001;30(no. 1):78–83.
[45] Lenhard, B., Hayes, W.S., Wasserman, W.W. GeneLynx: A Gene-Centric Portal to the Human Genome. Genome Research. 2001;11(no. 12):2151–2157.