P/FDM Mediator for a Bioinformatics Database Federation
The Internet is an increasingly important research tool for scientists working in biotechnology and the biological sciences. Many collections of biological data can be accessed via the World Wide Web, including data on protein and genome sequences and structure, expression data, biological pathways, and molecular interactions. Scientists’ ability to use these data resources effectively to explore hypotheses in silico is enhanced if it is easy to ask precise and complex questions that span across several different kinds of data resources to find the answer.
Some online data resources provide search facilities to enable scientists to find items of interest in a particular database more easily. However, working interactively with an Internet browser is extremely limited when one want to ask complex questions involving related data held at different locations and in different formats as one must formulate a series of data access requests, run these against the various databanks and databases, and then combine the results retrieved from the different sources. This is both awkward and time-consuming for the user.
To streamline this process, a federated architecture and the P/FDM Mediator are developed to integrate access to heterogeneous, distributed biological databases. The spectrum of choices for data integration is summarized in Figure 9.1. As advocated by Robbins, the approach presented in this chapter does not require that a common hardware platform or vendor database management system (DBMS) [1] be adopted by the participating sites. The approach presented here needs a “shared data model across participating sites,” but does not require that the participating sites all use the same data model internally. Rather, it is sufficient for the mediator to hold descriptions of the participating sites that are expressed in a common data model; in this system, the P/FDM Mediator, the functional data model [2] is used for this purpose. Tasks performed by the P/FDM Mediator include determining which external databases are relevant in answering users’ queries, dividing queries into parts that will be sent to different external databases, translating these subqueries into the language(s) of the external databases, and combining the results for presentation.

FIGURE 9.1 Continuum from tightly coupled to loosely coupled distributed systems involving multiple databases [1].
9.1 APPROACH
9.1.1 Alternative Architectures for Integrating Databases
The aim is to develop a system that will provide uniform access to heterogeneous databases via a single high-level query language or graphical interface and will enable multi-database queries. This objective is illustrated in Figure 9.2. Data replication and multi-databases are two alternative approaches that could help to meet this objective.

FIGURE 9.2 Users should be able to access heterogeneous, distributed bioinformatics resources via a single query language or graphical user interface.
Data Replication Approach
In a data replication architecture, all data from the various databases and databanks of interest would be copied to a single local data repository, under a single DBMS. This approach is taken by Rieche and Dittrich [3], who propose an architecture in which the contents of biological databanks including the EMBL nucleotide sequence databank and Swiss-Prot are imported into a central repository.
However, a data replication approach may not be appropriate for this application domain for several reasons. Significantly, by adopting a data repository approach, the advantages of the individual heterogeneous systems are lost. For example, many biological data resources have their own customized search capabilities tailored to the particular physical representation that best suits that data set. Rieche and Dittrich [3] acknowledge the need to use existing software and propose implementing exporters to export and convert data from the data repository into files that can be used as input to software tools.
Another disadvantage of a data replication approach is the time and effort required to maintain an up-to-date repository. Scientists want access to the most recent data as soon as they have been deposited in a databank. Therefore, whenever one of the contributing databases is updated, the same update should be made to the data repository.
Multi-Database Approach
A multi-database approach that makes use of existing remote data sources, with data described in terms of entities, attributes, and relationships in a high-level schema is favored. The schema is designed without regard to the physical storage format(s). Queries are expressed in terms of the conceptual schema, and it is the role of a complex software component called a mediator [4] to decide what component data sources need to be accessed to answer a particular query, organize the computation, and combine the results. Robbins [1] and Karp [5] have also advocated a federated, multi-database approach.
In contrast to a data replication approach, a multi-database approach takes advantage of the customized search capabilities of the component data sources in the federation by sending requests to these from the mediator. The component resources keep their autonomy, and users can continue to use them exactly as before. There is no local mirroring, and updates to the remote component databases are available immediately. A multi-database approach does not require that large data sets be imported from a variety of sources, and it is not necessary to convert all data for use with a single physical storage schema. However, extra effort is needed to achieve a mapping from the component databases onto the conceptual model.
9.1.2 The Functional Data Model
The system described in this chapter is based on the P/FDM object database system [6], which has been developed for storing and integrating protein structure data. P/FDM is itself based on the functional data model (FDM) [2], whose basic concepts are entities and functions. Entities are used to represent conceptual objects, while functions represent the properties of an object. Functions are used to model both scalar attributes, such as a protein structure’s resolution and the number of amino acid residues in a protein chain, and relationships, such as the relationship between chains and the residues they contain. Functions may be single-valued or multi-valued, and their values can either be stored or computed on demand. Entity classes can be arranged in subtype hierarchies, with subclasses inheriting the properties of their superclass, as well as having their own specialized properties. Contrast this with the more widely used relational data model whose basic concept is the relation—a rectangular table of data. Unlike the FDM, the relational data model does not directly support class hierarchies or many-to-many relationships.
Daplex is the query language associated with the FDM and, to illustrate the syntax of the language, Figure 9.3 shows two Daplex queries expressed against an antibody database [7]. Query A prints “the names of the amino acid residues found at the position identified by kabat code number 88 in variable domains of antibody light chains (VL domains).” This residue is located in the core of the VL domain and is spatially adjacent to the residue at Kabat position 23. Query B prints “the names of the residues at these two positions together with the computed distance between the centers of their alpha-carbon (CA) atoms.” Thus, one can explore a structural hypothesis about the spatial separation of these residues being related to the residue types occurring at these positions. In Query A, ig_domain is an entity class representing immunoglobulin domains, and the values that the variable d takes are the object identifiers of instances of that class. Domain_type and chain_type are string-valued functions defined on the class ig_domain. Domain_structure is a relationship function that returns the object identifier of the instance of the class structure that contains the ig_domain instance identified by the value of d. The expression protein_code (domain_structure (d)) illustrates an example of function composition. Query B shows nested loops in Daplex.

FIGURE 9.3 Daplex queries against an antibody database. Query A: “For each light chain variable domain, print the PDB entry code, the domain name, and the name of the residue occurring at Kabat position 88.” Query B: “For each VL domain, print the PDB entry code, the names of the residues at Kabat positions 23 and 88, and the distance between their alpha-carbon atoms.”
FDM had its origins in early work [2, 8], done before relational databases were a commercial product and before object-oriented programming (OOP) and windows, icons, menus, and pointers (WIMP) interfaces had caught on. Although it is an old model, it has adapted well to developments in computing because it was based on good principles. First, it was based on the use of values denoting persistent identifiers for instances of entity classes, as noted by Kulkarni and Atkinson [9]. This later became central to the object database manifesto [10]. Also, it had the notion of a subtype hierarchy, and it was not difficult to adapt this to include methods with overriding, as in OOP [11]. Second, the notions of an entity, a property, and a relationship (as represented by a function and its inverse) corresponded closely to the entity-relationship (ER) model and ER diagrams, which have stood the test of time. Third, it used a query language based on applicative expressions, which combined data extraction with computation. Thus, it was a mathematically well-formed language, based on the functional languages [12, 13], and it avoided the syntactic oddities of structured query language (SQL).
In developing the language since early work on the excluded function data model (EFDM) [9] Prolog was used as the implementation language because it is so good for pattern matching, program transformation, and code generation. Also, the data independence of the FDM, with its original roots in Multibase [14], allows to interface to a variety of kinds of data storage, instead of using a persistent programming language with its own data storage. Thus, unlike the relational or object-relational models, FDM does not have a particular notion of storage (row or tuple) built into it. Nor does it have fine details of arrays or record structures, as used in programming languages. Instead, it uses a mathematical notion of mapping from entity identifier to associated objects or values. Another change has been to strengthen the referential transparency of the original Daplex language by making it correspond more closely to Zermelo-Fraenkel set expressions (ZF-expressions), a name taken from the Miranda functional language [13, 15], and also called list comprehensions.
9.1.3 Schemas in the Federation
The design philosophy of P/FDM Mediator can be illustrated with reference to the three-schema architecture proposed by the ANSI Standards Planning And Requirements Committee (SPARC) [16]. This consists of the internal level, which describes the physical structure of the database; the conceptual level, which describes the database at a higher level and hides details of the physical storage; and the external level, which includes a number of external schemas or user views. This three-schema architecture promotes data independence by demanding that database systems be constructed so they provide both logical and physical data independence. Logical data independence provides that the conceptual data model must be able to evolve without changing external application programs. Only view definitions and mappings may need changing (e.g., to replace access to a stored field by access to a derived field calculated from others in the revised schema). Physical data independence allows to refine the internal schema for improved performance without needing to alter the way queries are formulated.
The clear separation between schemas at different levels helps in building a database federation in a modular fashion. In Figure 9.4 the ANSI-SPARC three-schema architecture is shown in two situations: in each of the individual data resources and in the mediator itself.

FIGURE 9.4 ANSI-SPARC schema architecture describing the mediator (left) and an external data resource (right).
First, consider an external data resource. The resource’s conceptual schema (which is called CR) describes the logical structure of the data contained in that resource. If the resource is a relational database, it will include information about table names, column names, and type information about stored values. With SRS [17], it is the databank names and field names. These systems also provide a mechanism for querying the data resource in terms of the table/class/databank names and column/tag/attribute/field names that are presented in the conceptual schema.
The internal schema (or storage schema, which is called IR) contains details of allocation of data records to storage areas, placement strategy, use of indexes, set ordering, and internal data structures that impact efficiency and implementation details [6]. This chapter is not concerned with the internal schemas of individual data resources. The mapping from the conceptual schema to the internal schema has already been implemented by others within each of the individual resources and it is assumed this has been done to make best use of the resources’ internal organization.
A resource’s external schema (which is called ER) describes a view onto the data resource’s conceptual schema. At its simplest, the external schema could be identical to the conceptual schema. However, the ANSI-SPARC model allows for differences between the schemas at these layers so different users and application programmers can each be presented with a view that best suits their individual requirements and access privileges. Thus, there can be many external schemas, each providing users with a different view onto the resource’s conceptual schema. A resource’s external, conceptual, and internal schemas are represented on the right side of Figure 9.4.
The ANSI-SPARC three-layer model can also be used to describe the mediator central to the database federation, and this is shown on the left side of Figure 9.4. The mediator’s conceptual schema (CM), also referred to as the federation’s integration schema, describes the content of the (virtual) data resources that are members of the federation, including the semantic relationships that hold between data items in these resources. This schema is expressed using the FDM because it makes computed data in a virtual resource. Both the derived results of arithmetic expressions and derived relationships look no different from stored data. Both are the result of functions—one calculates and the other extracts from storage. As far as possible, the CM is designed based on the semantics of the domain, rather than consideration of the actual partitioning and organization of data in the external resources. Thus, through functional mappings, different attributes of the same conceptual entity can be spread across different external data resources, and subclass–superclass relationships between entities in the conceptual model of the domain might not be present explicitly in the external resources [18].
No one can expect scientists to agree on a single schema. Different scientists are interested in different aspects of the data and will want to see data structured in a way that matches the concepts, attributes, and relationships in their own personal model. This is made possible by following the ANSI-SPARC model; the principle of logical data independence means the system can provide different users with different views onto the integration schema. EM is used to refer to an external schema presented to a user of the mediator. In this chapter, queries are expressed directly against an integration schema (CM), but these could alternatively be expressed against an external schema (EM). If so, an additional layer of mapping functions would be required to translate the query from EM to CM.
A vital task performed by the mediator is to map between CM and the union of the different CR. To facilitate this process, another schema layer that, in contrast to CM, is based on the structure and content of the external data resources is introduced. This schema is internal to the mediator and is referred to as IM. The mediator needs to have a view onto the data resource that matches this internal schema; thus, IM and ER should be the same. FDM is used to represent IM/ER. Having the same data model for CM and IM/ER brings advantages in processing multi-database queries, as will be seen in Section 9.1.4.
By redrawing Figure 9.4, the situation where there are u different external schemas presented to users and r data resources, the relationship between schemas in the federation is as shown in Figure 9.5. The five-level schema shown there is similar to that described by Sheth and Larson [19].
From past experience in building the prototype system, designing the IM/ER schema in a way that most directly describes the structure of a particular external resource adds practical benefits. In adding a remote resource to CM, one may focus mainly on those attributes in the remote resource that are important for making joins across to other resources. This clarifies the role of each schema level and the purpose of the query transformation task in transforming queries from one schema level to the next level down.
9.1.4 Mediator Architecture
The role of the mediator is to process queries expressed against the federation’s integration schema (CM). The mediator holds meta-data describing the integration schema and also the external schemas of each of the federation’s data resources (ER). In P/FDM, these meta-data are held, for convenience of pattern matching, as Prolog clauses compiled from high-level schema descriptions.
The architecture of the P/FDM Mediator is shown in Figure 9.6. The main components of the mediator are described in the following paragraphs.
The parser module reads a Daplex query (Daplex is the query language for the FDM), checks it for consistency against a schema (in this case the integration schema), and produces a list comprehension containing the essential elements of the query in a form that is easier to process than Daplex text (this internal form is called ICode).
The simplifier’s role is to produce shorter, more elegant, and more consistent ICode, mainly through removing redundant variables and expressions (e.g., if the ICode contains an expression equating two variables, that expression can be eliminated, provided that all references to one variable are replaced by references to the other), and flattening out nested expressions where this does not change the meaning of the query. Essentially, simplifying the ICode form of a query makes the subsequent query processing steps more efficient by reducing the number of equivalent ICode combinations that need to be checked.
The rule-based rewriter matches expressions in the query with patterns present on the left-hand side of declarative rewrite rules and replaces these with the right-hand side of the rewrite rule after making appropriate variable substitutions. Rewrite rules can be used to perform semantic query optimization. This capability is important because graphical interfaces make it easy for users to express inefficient queries that cannot always be optimized using general purpose query optimization strategies. This is because transforming the original query to a more efficient one may require domain knowledge (e.g., two or more alternative navigation paths may exist between distantly related object classes but domain knowledge is needed to recognize that these are indeed equivalent).
A recent enhancement to the mediator is an extension to the Daplex compiler that allows generic rewrite rules to be expressed using a declarative high-level syntax [20]. This makes it easy to add new query optimization strategies to the mediator.
The optimizer module performs generic query optimization.
The reordering module reorders expressions in the ICode to ensure that all variable dependencies are observed.
The condition compiler reads declarative statements about conditions that must hold between data items in different external data resources so these values can be mapped onto the integration schema.
The ICode rewriter expands the original ICode by applying mapping functions that transform references to the integration schema into references to the federation’s component databases. Essentially the same rewriter mentioned previously is used here, but with a different set of rewrite rules. These rewrite rules enhance the ICode by adding tags to indicate the actual data sources that contain particular entity classes and attribute values. Thus, the ICode rewriter transforms the query expressed against the CM into a query expressed against the ER of one or more external databases.
The crucial idea behind the query splitter is to move selective filter operations in the query down into the appropriate chunks so they can be applied early and efficiently using local search facilities as registered with the mediator [KIG94]. The mediator identifies which external databases hold data referred to by parts of an integrated query by inspecting the meta-data, and adjacent query elements referring to the same database are grouped together into chunks. Query chunks are shuffled and variable dependencies are checked to produce alternative execution plans. A generic description of costs is used to select a good schedule/sequence of instructions for accessing the remote databases.
Each ICode chunk is sent to one of several code generators. These translate ICode into queries that are executable by the remote databases, transforming query fragments from ER to CR. New code generators can be linked into the mediator at runtime.
Wrappers deal with communication with the external data resources. They consist of two parts: code responsible for sending queries to remote resources and code that receives and parses the results returned from the remote resources. Wrappers for new resources can be linked into the mediator at runtime. Note that a wrapper can only make use of whatever querying facilities are provided by the federation’s component databases. Thus, the mediator’s conceptual model (CM) will only be able to map onto those data values that are identified in the remote resource’s conceptual model (CR). Thus, queries involving concepts like gene and chromosome in CM can only be transformed into queries that run against a remote resource if that resource exports these concepts.
The result fuser provides a synchronization layer, which combines results retrieved from external databases so the rest of the query can proceed smoothly. The result fuser interacts tightly with the wrappers.
9.1.5 Example
A prototype mediator has been used to combine access databanks at the EBI via an SRS server [17] and (remote) P/FDM test servers. Remote access to a P/FDM database is provided through a CORBA server [21]. This example, using a small integration schema, illustrates the steps involved in processing multi-database queries.
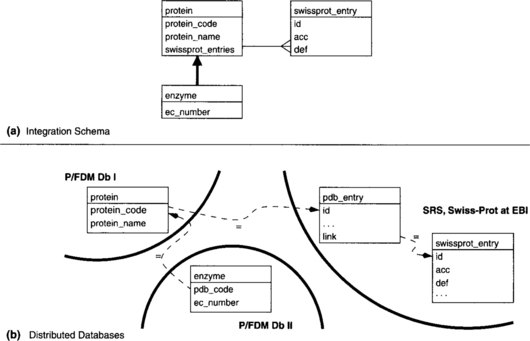
In this example, three different databases are viewed through a unifying integration schema (CM), which is shown in Figure 9.7(a). There are three classes in this schema: protein, enzyme, and swissprot_entry. A function representing the enzyme classification number (ec_number) is defined on the class enzyme, and enzymes inherit those functions that are declared on the superclass protein. Each instance of the class protein can be related to a set of swissprot_entry instances.
Figure 9.7(b) shows the actual distribution of data across the three databases; each of these three databases has its own external schema, ER. Db I is a P/FDM database that contains the codes and name of proteins. Db II is also a P/FDM database and contains the protein code (here called pdb-code) and enzyme classification code of enzymes. To identify Swiss-Prot entries at the EBI that are related to a given protein instance, one must first identify the Protein Data Bank (PDB) entry whose ID matches the protein code and then follow further links to find related Swiss-Prot entries. Relationships between data in remote databases can be defined by conditions that must hold between the values of the related objects. Constraints on identifying values are represented by dashed arrows in Figure 9.7.
Figure 9.8 shows a Daplex query expressed against the integration schema, CM. This query prints information about enzymes that satisfy certain selection criteria and their related Swiss-Prot entries. Figure 9.9 shows a pretty-printed version of the ICode produced when this query is compiled. This ICode is then processed by the query splitter, producing ICode (in terms of the resources’ external schemas, ER) that will be turned into queries that will be sent to the three external data resources (Figure 9.10). Note that the variable V1 is common to all three query fragments. Values for this variable retrieved from P/FDM Db II are used in constructing queries to be sent to the other data resources.

FIGURE 9.9 ICode corresponding to the query in Figure 9.8.

FIGURE 9.10 ICode sub-queries against the actual data resources that need to be accessed to answer the query in Figure 9.8.
In the example, the class protein is related to the class swissprot_entry in the integration schema by a multi-valued relationship function called swissprot_entries. The mapping function given in Figure 9.11 is used in transforming queries that contain this relationship into enhanced ICode that refers to the external schemas, ER, of the actual data resources. Mapping functions such as this can be compiled from high-level declarative rewrite rules and do not have to be written by hand.
9.1.6 Query Capabilities
Daplex is the query language of the system. The examples in Figure 9.3 and Figure 9.12 show how function calls can be composed in queries. The compositional form makes it easy to write complex queries and computations over the database that can be optimized by a query optimizer. This is a point often overlooked by the OOP community; Java and C++ have the necessary expressiveness, but because they lack referential transparency and a data model it is hard to make general optimizers for database applications in them. Daplex has greater expressive power that SQL (e.g., recursive functions can be defined directly in Daplex). This is particularly useful in many areas of bioinformatics, such as following transitive relationships through a sequence of reactions in a biochemical pathway or finding related biological terms in a hierarchical vocabulary.
As mentioned in Section 9.1.4, Daplex queries are converted into ICode for subsequent processing. The great advantage is that many important optimizations just involve reordering selection predicates and generators in the set expression. These, in turn, are conveniently implemented as rewrite rules in Prolog [6]. It was also shown how to expand definitions of derived functions in the course of optimizing set expressions [22]. This makes good use of the referential transparency of expressions in functional programming. By contrast, where the computation is embedded in C++ or Visual Basic with arbitrary assignments, it is very hard to do significant optimization. This has led to the widespread adoption of comprehensions (as ZF-expressions are now called). Buneman et al. [23] have shown the importance of distinguishing list, bag, and set comprehensions, so, strictly speaking, ZF-expressions compute bags but represent them by lists.
The Daplex query language enables arbitrary calculations to be combined with data retrieval operations [7]. For example, Figure 9.12 shows a query, expressed against an integration schema, that performs a geometric calculation on data in an antibody database and relates objects satisfying the given criteria with data in a remote Swiss-Prot database. The function distance computes a value rather than retrieving a stored value. As explained in Section 9.1.2, functions whose values are stored or derived look the same in the query, and the user cannot tell from looking at a result value whether it was retrieved from disc or computed.
Daplex queries frequently include calls to functions written in procedural languages. For example, when working with data on 3D protein structures, one often calls out to geometric code from within queries, including C routines to measure bond angles and torsion angles, and code to superpose one structural fragment on another to compare 3D similarity. Following the same approach, it would be possible to treat the results of a computation on a remote machine, such as a BLAST search, that are generated dynamically at run-time just like data values that are stored persistently on disc. The system does not yet have a wrapper for BLAST, but, in principle, such a wrapper would be implemented just like any other derived function in P/FDM.
The mediator does not currently cache query results, so subsequent queries cannot refer to a result set. However, both user interfaces described in Section 9.2.2 enable follow-on queries to be constructed incrementally based on the previous query.
For more complex P/FDM applications that cannot be expressed in Daplex, Prolog can be used [24]. However, unlike Daplex queries, these Prolog programs are not optimized automatically (see Section 9.2.1). The P/FDM system provides a set of Prolog routines that perform primitive data access operations, such as retrieving the object identifier of an instance of an entity class, retrieving the scalar value of an attribute of an object with a given object identifier, or retrieving the object identifier of a related object. Queries that require access to several data sources in the federation can be written directly in Prolog.
9.1.7 Data Sources
P/FDM was previously used with various data sources including hash files, relational databases, flat files (including some accessed via SRS), POET, AMOS II and AceDB.
There is no control over changes being made to remote resources, as remote sites retain their autonomy. Depending on the nature of the changes to a remote resource, this may require changes in one or both parts of the wrapper for that resource. Changes to a remote resource need not require changes to be made to the mediator’s conceptual model (CM), though they may require some changes to be made to the declarative mapping functions associated with data in the changed resource.
9.2 ANALYSIS
The use of mediators was originally proposed by Wiederhold [4] and became an important part of the knowledge sharing effort architecture [25]. Examples of such intelligent, information-seeking architectures are Infosleuth [26] and KRAFT [27]. In this architecture, the mediator can run on the client machine, or else be available as middleware on some shared machine, while the wrapper is on the remote machine containing the knowledge source. The idea behind this is that existing knowledge sources can evolve their schemas, yet present a consistent interface to the mediator via suitable changes to the wrapper. For this purpose the wrapper may be as simple as an SQL view, or it may be more complex, involving mapping of code. In any case, the site is able to preserve some local autonomy. Other mediators do not have to worry about how the site evolves internally. Also, new sites can join a growing network by registering themselves with a facilitator. All the mediator needs to know is how to contact the facilitator and that any knowledge sources the facilitator recommends will conform to the integration schema.
This chapter describes an alternative architecture, where the wrappers reside with the mediator. This has the advantage that there is no need to get the knowledge source to install and maintain custom-provided wrapper software.
In the architecture, shown in Figure 9.6, the code generators produce code in a different query language or constraint language. Thus, they are used in two directions. In one direction, they map queries or constraints into a language that can be used directly at the knowledge source. This can be crucial for efficiency because it allows one to move selection predicates closer to the knowledge source in a form that is capable of using local indexes. This can have a significant effect with database queries because it saves bringing many penny packets of data back through the interface, only to be filtered and rejected on the far side [28]. In the other direction, wrappers are used to map data values (e.g., by using scaling factors to change units or by using a lookup table to replace values by their new identifiers).
Note that building a so-called global integration schema is not advocated. These have often been criticized on the grounds that attempts to map every single concept in one all-embracing schema is both laborious and never-ending. Instead, an incrementally growing integration schema is visualized, driven by user needs. Ideally the schema would be built interactively using a GUI and rules that suggest various mappings, as proposed by Mitra et al. [29] in their ONION system for incremental development of ontology mappings. The crucial thing to realize is that the integration schema represents a virtual database, which allows it to evolve much more easily than a physical database.
Related work in the bioinformatics field includes the Kleisli system presented in Chapter 6 [30, 31]. The query language used in Kleisli is the Collection Programming Language (CPL), which is a comprehension-based language in which the generators are calls to library functions that request data from specific databases according to specific criteria. Thus, when writing queries, the user must be aware of how data are partitioned across external sites. This contrasts with the approach taken in the P/FDM Mediator, where references to particular resources do not feature in the integration schema or in user queries. Of course, an interface based on domain concepts and without references to particular resources could be built on top of Kleisli.
The TAMBIS system presented in Chapter 7 [32] writes query plans in CPL. Plans in TAMBIS are based on a classification hierarchy, whereas P/FDM plans are oriented toward ad hoc SQL3-like queries. However, the overall approach is similar to using a high-level intermediate code translated through wrappers.
Another related project is DiscoveryLink, presented in Chapter 11 [33]. The architecture of the DiscoveryLink system is similar to that presented in this chapter. DiscoveryLink uses the relational data model instead of FDM, and all the databases accessed via DiscoveryLink must present an SQL interface.
9.2.1 Optimization
Optimization takes a great advantage from using an easily transformable high-level representation based on functional composition.
Three kinds of optimization are done within the P/FDM Mediator. First, the rewriter can apply rules that perform semantic query optimization. Additionally, rewrite rules [20] can be used to implement the logical rules given by Jarke and Koch [34]. They can implement many forms of rewrites based on data semantics, as discussed in King [35]. They can spot opportunities to replace iteration by indexed search [33]. In experiments using the AMOS II system [36] as a remote resource, rewrite rules were able to implement flattening and un-nesting transformations that prevent wasting time compiling subqueries in AMOSQL. A similar approach could be adapted to features of other DBMSs. Most importantly, rewrites that change the relative workload between two processors in a distributed query can be performed. Finally, all these rewrites can be combined, as some of them will enable others to take place. Thus, one can deal with many combinations without having to foresee them and code them individually.
Second, the optimizer performs generic query optimizations. The philosophy of the optimizer is to use heuristics to improve queries. It examines alternative execution plans and, although it uses a simple cost model, it is successful in avoiding inefficient strategies, and it often selects the most effective approach [15]. The optimizer was subsequently rewritten using a simple heuristic to avoid the combinatorial problem of examining all possible execution plans for complex queries [22].
Third, the query splitter attempts to group together query elements into chunks that can be sent as single units to the external data resources, thus providing the remote system with as much information as possible to give it greater scope for optimizing the sub-query.
Outside the mediator, the approach is able to take advantage of the optimization capabilities of the external resources.
There is scope for introducing adaptive query processing techniques to improve the execution plans as execution proceeds and as results are returned to the mediator [37], but this has not yet been done in our prototype system.
9.2.2 User Interfaces
While queries against a P/FDM schema can be formulated directly in either Prolog or Daplex, this requires some programming ability and care must be taken to use the correct syntax. Therefore, two interfaces were developed to formulate queries without the user having to learn to program in either Prolog or Daplex. Both of these interfaces have a representation of the schema at their heart, and they enable the user to construct well-formed Daplex queries by clicking and typing values for attributes to restrict the result set.
A Java-based visual interface for P/FDM [38] was developed with a graphical representation of the database schema at its center. Figure 9.13 shows this interface in use with the schema for an antibody database [7]. Users construct queries by clicking on entity classes and relationships in the schema diagram and constraining the values of attributes selected from menus. As this is done, the Daplex text of the query under construction is built up in a sub-window (the query editor window). Queries are submitted to the database via a CORBA interface [21]. Results satisfying the selection criteria are displayed in a table in a separate result window. A particularly novel feature of the interface is copy-and-drop, which enables the user to select and copy data values in the result window and then drop these into the query editor window. When this is done, the selected values are merged into the original query automatically, in the appropriate place in the query text, to produce a more specialized query. The Java-based interface runs as a Java application, but it does not yet run within a Web browser.

FIGURE 9.13 Visual Navigator query interface [38].
In addition, a Web interface was developed with hypertext markup language (HTML) forms and accesses the mediator via a CGI program (Figure 9.14). Such interfaces can be generated automatically from a schema file. The interface’s front page lists the entity classes in the schema, and the user selects one of these as the starting point for the query. As the query is built up, checkboxes are used to indicate those attributes whose values are to be printed. The user can constrain the value of an attribute by typing into its entry box (e.g., 1.1.1.1 or <2.5) and can navigate to related objects using the selection box labeled relationships at the bottom of each object’s representation within the Web page. Figure 9.14 shows the Web interface at the point where the user has formulated the query used in the example in Section 9.1.5. Pressing the Submit button will cause the equivalent Daplex query to be generated.
When using a graphical user interface that supports ad hoc querying, it is easy for naive queries that involve little or no data filtering to be expressed. This can result in queries that request huge result sets from remote resources. An alternative approach, as in TAMBIS, would be to provide only user interfaces that guide the user toward constructing queries with a particular structure and that have a suitable degree of filtering. However, such an interface would constrain the user to asking only parameterized variants of a set of canned queries anticipated by the interface designer. While such interfaces could be implemented easily, P/FDM design specification favors that users have the freedom to express arbitrary queries against a schema, and an area for future work is identifying and dealing with queries that could place unreasonable loads on the component databases in the federation.
Current interfaces do not provide personalization capabilities. It is, however, possible to provide users with their individual views of the federation (see EM schemas in Figure 9.5), but this would be done by the database federation’s administrator on behalf of users, rather than by users themselves.
9.2.3 Scalability
When a new external resource is added to the federation, the contents of that resource must be described in terms of entities, attributes, and relationships—the basic concepts in the FDM. For example, entity classes and attributes are used to describe the tables and columns in a relational database, the classes and tags in an AceDB database, and the databanks and fields accessed by SRS. The integration schema has to be extended to include concepts in the new resource, and mapping functions to be used by the ICode rewriter must be generated. Because the mediator has a modular architecture in which query transformation is done in stages, the only new software components that might have to be written are code generators and wrappers—the components shown with dark borders in Figure 9.6. All other components within the mediator are generic. However, the federation administrator might want to add declarative rewrite rules that can be used by the rewriter to improve the performance of queries involving the new resource. Code generators for new data sources can be written in one or two days when using existing code generators as a guide. In general, as the expressions being evaluated obey the principles of substitutability and referential integrity, expressions that match patterns in rewrite rules can be substituted with other expressions that have the same value. This means new mappings can be added without the risk of encountering special cases or some arbitrary limit on the complexity of expressions, as can happen with SQL.
A federated database system and a mediator system are similar architectures that differ in terms of how easily one can attach new database sources. In a federated architecture, the integration schema is relatively fixed and designed with particular database sources in mind. Extra databases can be added, with some effort, by the database administrator. A mediator tries to integrate new databases available at their given Web addresses on the basis of descriptions provided by the end user. The whole process is more dynamic. When dealing with a new source, a good mediator will try to spot heuristic optimization rules it can re-use from similar databases it knows about. In general, it is more intelligent and less reliant on human intervention. A long-term goal is that, as a suite of code generators and wrappers is added to the P/FDM Mediator, it will become easy to add new resources by presenting the mediator with new remote schemas and specifying which code generators and wrappers should be used.
9.3 CONCLUSIONS
The P/FDM Mediator is a computer program that supports transparent and integrated access to different data collections and resources. Ad hoc queries can be asked against an integration schema, which is a pre-defined collection of entity classes, attributes, and relationships. The integration schema can be extended at any time by adding declarative descriptions of new data resources to the mediator’s set-up files.
Rather than building a data warehouse, the developed system brings data from remote sites on demand. The P/FDM Mediator arranges for this to happen without further human intervention. The presented approach preserves the autonomy of the external data resources and makes use of existing search capabilities implemented in those systems.
Bioinformatics faces a “crisis of data integration” [1], which is best addressed through federations that allow their constituent databases to develop autonomously and independently. The existence of schemas at different levels, as shown in Section 9.1.3, makes apparent the requirements for query transformation in a mediator in a database federation. The transformations in the system are all based on well-defined mathematical theory using function composition, as pioneered by Shipman [2] and Buneman [12]. This results in a modular design for the mediator that enables the federation to evolve incrementally.
ACKNOWLEDGMENT
The prototype P/FDM Mediator described in this chapter was implemented by Nicos Angelopoulos. This work was supported by a grant from the BBSRC/EPSRC Joint Programme in Bioinformatics (Grant Ref. 1/BIF06716).
REFERENCES
[1] Robbins, R.J. Bioinformatics: Essential Infrastructure for Global Biology. Journal of Computational Biology. 1996;3(no. 3):465–478.
[2] Shipman, D.W. The Functional Data Model and the Data Language DAPLEX. ACM Transactions on Database Systems. 1981;6(no. 1):140–173.
[3] Rieche, B., Dittrich, K.R. A Federated DBMS-Based Integrated Environment for Molecular Biology. In: In Proceedings of the Seventh International Conference on Scientific and Statistical Database Management. Los Alamitos, CA: IEEE Computer Society; 1994:118–127.
[4] Wiederhold, G. Mediators in the Architecture of Future Information Systems. IEEE Computer. 1992;25(no. 3):38–49.
[5] Karp, P.D., A Vision of DB Interoperation, Proceedings of the Second Meeting on the Interconnection of Molecular Biology Databases. Cambridge, UK. July. 1995:20–22.
[6] Gray, P.M.D., Kulkarni, K.G., Paton, N.W. Object-Oriented Databases: A Semantic Data Model Approach. Hemel Hempstead, Hertfordshire: Prentice Hall International; 1992.
[7] Kemp, G.J.L., Jiao, Z., Gray, P.M.D., et al. Combining Computation with Database Accesss in Biomolecular Computing. In: Litwin W., Risch T., eds. Applications of Databases: Proceedings of the First International Conference. Heidelberg, Germany: Springer-Verlag; 1994:317–335.
[8] Kerschberg, L., Pacheco, J.E.S. A Functional Data Model. Rio de Janeiro, Brazil: Department of Informatics, Universidade Catolica Rio de Janeiro; 1976.
[9] Kulkarni, K.G., Atkinson, M.P. EDFM: Extended Functional Data Model. The Computer Journal. 1986;29(no. 1):38–46.
[10] Atkinson, M., Bancilhon, F., DeWitt, D., et al. Deductive and Object-Oriented Databases. In: Kim W., Nicholas J-M., Nishio S., eds. Proceedings of the 1st International Conference on Deductive and Object-Oriented Databases (DOOD ’89). Amsterdam, The Netherlands: North-Holland; 1990:223–240.
[11] P.M.D., Gray, D.S., Moffat, N.W., Paton. A Prolog Interface to a Functional Data Model Database. In: Smith J.W., Ceri S., Missikoff M., eds. Advances in Database Technolog3-EDBT ’88. Heidelberg, Germany: Springer-Verlag; 1988:34–48.
[12] Buneman, P., Frankel, R.E. Fql: A Functional Query Language. In: Bernstein P.A., ed. Proceedings of the ACM SIGMOD International Conference on Management of Data. Boston: ACM Press; 1979:52–58.
[13] Turner, D.A., Miranda: A Non-Strict Functional Language with Polymorphic Types. Jouannaud J-P., ed. Functional Programming Languages and Computer Architecture, Lecture Notes in Computing Science. Heidelberg, Germany: Springer-Verlag; 1985;Vol. 201:1–16.
[14] Landers, T.A., Rosenberg, R.L. An Overview of MULTIBASE. In: Schneider H-J., ed. Distributed Data Bases, Proceedings of the 2nd International Symposium on Distributed Data Bases. Amsterdam, The Netherlands: North-Holland; 1982:153–184.
[15] Paton, N.W., Gray, P.M.D. Optimising and Executing Daplex Queries Using Prolog. The Computer Journal. 1990;33(no. 6):547–555.
[16] ANSI Standards Planning and Requirements Committee. Interim Report of the ANSI/X3/SPARC Study Group on Data Base Management Systems. FDT—Bulletin of ACM SIGMOD. 1975;7(no. 2):1–140.
[17] Ezold, T., Argos, P. SRS Indexing and Retrieval Tool for Flat File Data Libraries. Computer Applications in the Biosciences. 1993;9(no. 1):49–57.
[18] Grufman, S., Samson, F., Embury, S.M., et al. Distributing Semantic Constraints Between Heterogenous Databases. In: Gray A., Larson P-A., eds. Proceedings of the 13th Annual Conference on Data Engineering. New York: IEEE Computer Society Press, 1997.
[19] Sheth, A.P., Larson, J.A. Federated Database Systems for Managing Distributed, Heterogenous and Autonomous Databases. ACM Computing Surveys. 1990;22(no. 3):183–236.
[20] Kemp, G.J.L., Gray, P.M.D., Sjöstedt, A.R. Improving Federated Database Queries Using Declarative Rewrite Rules for Quantified Subqueries. Journal of Intelligent Information Systems. 2001;17(no.2–3):281–299.
[21] Kemp, G.J.L., Robertson, C.J., Gray, P.M.D., et al. CORBA and XML: Design Choices for Database Federations. In: Lings B, Jeffery K., eds. Proceedings of the Seventeenth British National Conference on Databases. Heidelberg, Germany: Springer-Verlag; 2000:191–208.
[22] Jiao, Z., Gray, P.M.D. Optimisation of Methods in a Navigational Query Language. In: Delobel C., Kifer M, Masunaga Y., eds. Proceedings of the Second International Conference on Deductive and Object-Oriented Databases. Heidelberg, Germany: Springer-Verlag; 1991:22–42.
[23] March Buneman, P., Libkin, L., Suciu, D., et al. Comprehension Syntax. SIGMOD Record. 1994;23(no. 1):87–96.
[24] Kemp, G.J.L., Gray, P.M.D. Finding Hydrophobie Microdomains Using an Object-Oriented Database. Computer Applications in the Biosciences. 1990;6(no. 4):357–399.
[25] Neches, R., Fikes, R., Finin, T., et al. Enabling Technology for Knowledge Sharing. Artificial Intelligence. 1991;12(no. 3):36–56.
[26] Bayardo, R.J., Bohrer, B., Brice, R.S., et al. InfoSleuth: Semantic Integration of Information in Open and Dynamic Environments. In: Peckham J., ed. SIGMOD 1997, Proceedings of the ACM SIGMOD International Conference on Very Large Data Bases. New York: ACM Press; 1997:195–206.
[27] Gray, P.M.D., Preece, A.D., Fiddian, N.J., et al. KRAFT: Knowledge Fusion from Distributed Databases and Knowledge Bases. In: Wagner R.R., ed. Proceedings of the 8th International Workshop on Database and Expert Systems Applications. Los Alamitos, CA: IEEE Computer Society Press; 1997:682–691.
[28] Kemp, G.J.L., Iriarte, J.J., Gray, P.M.D. Efficient Access to FDM Objects Stored in a Relational Database. In: Bowers D.S., ed. Directions in Databases: Proceedings of the Twelfth British National Conference on Databases. Heidelberg, Germany: Springer-Verlag; 1994:170–186.
[29] Mitra, P., Weiderhold, G., Kersten, M. A Graph-Oriented Model for Articulation of Ontology Interdependencies. In: Zaniolo C., Lockerman P.C., Scholl M.H., et al, eds. Advanced Database Technology—EDBT 2000. Heidelberg, Germany: Springer-Verlag; 2000:86–100.
[30] Buneman, P., Davidson, S.B., Hart, K., et al. A Data Transformation System for Biological Data Sources. In: Dayal U., Gray P.M.D., Nishio S., eds. VLDB ’95, Proceedings of the 21st International Conference on Very Large Data Bases. San Francisco: Morgan Kaufmann; 1995:158–169.
[31] Wong, L. Kleisli: Its Exchange Format, Supporting Tools and an Application in Protein Interaction Extraction. In: Proceedings of the IEEE International Symposium on Bio-Informatics and Biomedical Engineering. New York: IEEE Computer Society Press; 2000:21–28.
[32] Paton, N.W., Stevens, R., Baker, P., et al. Query Processing in the TAMBIS Bioinformatics Source Integration System. In: Proceedings of the 11th International Conference on Scientific and Statistical Database Management. New York: IEEE Computer Society Press; 1999:138–147.
[33] Haas, L.M., Kodali, P., Rice, J.E., et al. Integrating Life Sciences Data—With A Little Garlic. In: IEEE International Symposium on Bio-Informatics and Biomedical Engineering. New York: IEEE Computer Society Press; 2000:5–12.
[34] Jarke, M., Koch, J. Range Nesting: A Fast Method to Evaluate Quantified Queries. In: DeWitt D.J., Gandarin G., eds. SIGMOD ’83, Proceedings of the Annual Meeting. Boston: ACM Press; 1983:196–206.
[35] King, J.J. Query Optimisation by Semantic Reasoning. Ann Arbor, MI: University of Michigan Press; 1984.
[36] June 23 Risch, T., Josifovski, V., Katchaounov, T. AMOS II Concepts. 2000. http://www.dis.uu.se/∼udbllamosldoclamos.concepts.html
[37] Ives, Z.G., Florescu, D., Friedman, M., et al. An Adaptive Query Execution System for Data Integration. In: Delis A., Faloutsos C., Ghandeharizadeh S., eds. SIGMOD 1999, Proceedings of the ACM SIGMOD International Conference on Management of Data. Boston: ACM Press; 1999:299–310.
[38] Gil, I., Gray, P.M.D., Kemp, G.J.L. A Visual Interface and Navigator for the P/DFM Object Database. In: Paton N.W., Griffiths T., eds. 1999 User Interfaces to Data Intensive Systems. Los Alamitos, CA: IEEE Computer Society Press; 1999:54–63.