Chapter 13
Open Science Data Repository for Toxicology
Valery Tkachenko1, Richard Zakharov2 and Sean Ekins3
1Rockville, MD, USA
2Rockville, MD, USA
3Collaborations Pharmaceuticals, Inc., Raleigh, NC, USA
Chapter Menu
13.1 Introduction
Tools for collaboration predominantly revolve around desktop computer applications [1, 2] and use “software as a service” as a business model [1–3]. These desktop software have also become more widely accessible in academia and small companies (e.g., CDD Vault, Science Cloud). Such applications are useful for secure sharing of data with collaborators in which retention of intellectual property (IP) is important. However, we are increasingly seeing a shift to more companies, institutes, and researchers openly sharing data, regardless of IP. While this has been predominantly in the neglected disease space (GlaxoSmithKline, Novartis, and St. Jude's sharing malaria and tuberculosis data) [4–8] this is starting to broaden, for example, GlaxoSmithKline sharing of kinase data [9] and AstraZeneca sharing their Absorption, Distribution, Metabolism and Excretion (ADME) data on ChEMBL [10]. Alongside this, there are increasing efforts from researchers to publish in open access journals and release data into open or free databases [11, 12]. In addition, there are an increasing number of online tools that are viable for storing science-related content, for example, FigShare (Digital Science), SlideShare (Microsoft), Mendeley (Elsevier), Dropbox, and so on and consortia to coordinate and make data accessible, such as OpenPHACTS [13, 14], as well as a myriad of other initiatives to free up data [15–17]. There are also repositories such as Open Software Foundation (OSF), Dryad, Dataverse, DataHub, Kaggle, and others which may or may not owned be by publishers or other commercial organizations and some cases which are open source. This raises the question how can we possibly connect all this predominantly open data? Social media themselves can be useful for sharing data, such as blogs and wikis in which open scientists can describe their scientific methods and results, link to other content (uploaded images, graphs, and models), and beyond [11, 12]. We are also seeing the benefits of investments over the last decade in high-throughput screening that has resulted in large structure-activity datasets entering public and open databases. For example the openly available ChEMBL [10, 18] database has already been assembled by careful curation, and provides the necessary raw data to kick-start literally thousands of groups of structure–activity datasets, of various sizes. Furthermore, the massive PubChem database is frequently used by scientists to deposit raw data in its original, machine-readable form as an adjunct to publication. The EPA Tox21 measurements against hundreds of targets [19] represents a third large dataset. However, the quality of the data for some might still be questionable. While all these resources are great sources of information they are still prone to a series of fundamental weaknesses which were introduced by design and cannot be incrementally eliminated. We therefore envisage in future a massive growth in open science and we predict the need for an open science data repository (OSDR) which would be particularly valuable for sharing curated data in real time and making models accessible, including those based on toxicology data.
13.2 Open Science Data Repository
We believe an OSDR will be useful for connecting scientists and sharing data for many types of projects. This represents a potential knowledge base for real-time management of various scientific data. OSDR fully adopts FAIR data principles (data fully supporting the FAIR - findable, accessible, interoperable, reusable - principle for research data findable, accessible, interoperable, reusable [20]). All data in OSDR are assigned a globally unique identifier, besides which data parts can be assigned a Digital Object Identifier (DOI). All data are described with rich metadata. Data and metadata become immediately searchable on creation or update. Metadata exposed through RESTful Application Program Interface (API) conform through “maturity level 3” and contain data identifiers. Both data and metadata are accessible through RESTful API. In fact, the whole OSDR is built as a swarm of microservices communications among one another and with User Interface (UI) through RESTful APIs. Data and metadata layers in ODSP are separated and have various levels of persistence, guaranteeing that metadata remain accessible even when data are not available anymore. Both data and metadata that persist in OSDR are exposed through RESTful API endpoints supporting a standard (extensible) set of ontologies and standard formats and protocols. Metadata use controlled vocabularies where possible, but OSDR also supports a process of building custom-controlled vocabularies. RESTful API supports “maturity level 3.” The OSDR data model is organized as a set of arbitrary hierarchical entities with the ability to associate metadata documents with the necessary plurality at each level of hierarchy. All OSDR entities are associated with the license or the license can be derived from constituent parts. All entities are associated with their provenance pointing to the source of the data record. Data and metadata in OSDR are grouped into domain-specific and ontology-driven groupings. The technology therefore can be readily used for sharing toxicology data and models (similar to those described in other chapters of this book). We believe open sharing of data with OSDR will also facilitate scientific insights and the discovery of new therapeutic approaches when used for drug discovery applications. OSDR provides capabilities for depositing and managing general data, it has a unique architecture that allows to connect new chemistry-intelligent modules to the existing data processing and curation pipeline. To provide some perspective, some comparisons with other databases and repositories are discussed in the following.
PubChem [21] is a database built to deposit, process, and provide public access to chemical data, but its data-deposition pipeline is rigid and there are no real-time user curation capabilities. ChEMBL [10] and ChEBI [22] are highly curated chemical databases with heavy ontological information content, but without online curation. Figshare [23] is a general-purpose data repository with a heavy emphasis on scientific content and with one of its key features being the ability to generate and assign DOI to the data. Mendeley's [24] data repository provides similar capabilities to those of Figshare. OpenPHACTS [14] was proposed as a semantic web knowledge base to provide private/public access to a variety of chemical and biological information. In reality, integration of data from multiple data sources is so challenging that data updates can be made once every few months at best, rather than in real-time as was expected. OpenPHACTS has also provided very valuable outcomes which we have built on, namely, its (i) data collections (ii) theoretical basis (e.g., ontologies), and (iii) practical experience. Our goal therefore is to address all these deficiencies accumulated in other systems and make OSDR the de-facto gold standard for open science. Such a tool will then enable improved data sharing and collaboration with potential impact across many scientific areas.
The value proposition for the use of OSDR can be summarized as follows. The project is intended to bring together open and prepublication data and then facilitate research around the data. By connecting focused groups of disparate individuals and organizations scattered around the country or globe, ample opportunity exists to both gather and disseminate important information to a highly relevant target audience. This includes information about new scientific developments of interest. Team members will be able to borrow and reuse a growing collection of existing data. This should result in new technology which would benefit the toxicology, neglected, rare disease communities (as examples) as well as far beyond. We are making the OSDR open source [25], will provide support for the platform, offer custom development services, and license the API. This is a viable and well-established business model for open source software which will be leveraged.
13.3 Benefits of OSDR
We are pioneering key innovations in the prototype OSDR which will likely be of value to customers (Figure 13.1) with a broad array of features and applications (Figures 13.2–13.4).

Figure 13.1 (1) Examples of the OSDR prototype to date showing bidirectional integration with various cloud drives allows seamless data transfers between cloud storage and OSDR; (2) web user interface also allows intuitive data deposition using drag and drop; (3) concise filter system provides a convenient way of navigating information stored or indexed in OSDR; (4) hierarchical presentation of information allows one to arrange the data based on organization or research structure; (5) standard CMS (content management system) operations are supported; (6) various view modes allow representing complex information in a visual and concise manner; (7) user interface based on modern web frameworks provides an excellent user experience.

Figure 13.2 Examples of the OSDR prototype to date showing OSDR tabular data entry. Mapping columns from a CSV file (1) to their semantic meaning (2) allows to resolve entries in real-time into a set of public database identifiers (3, ChemSpider, ChEMBL, PubChem), create a chemical structure from provided information (4), and calculate conversion confidence value based on a set of mappings (e.g., chemical name, InChI, SMILES).


Figure 13.3 Examples of the OSDR prototype to date. (a) Document browse mode with thumbnail previews. (b) Document view mode with a larger preview and other information arranged into infoboxes.

Figure 13.4 Examples of the OSDR prototype to date. Built in preview mode showing different file types (1, word; 2, excel; 3, powerpoint; 4, PDF).
13.3.1 Chemically and Semantically Enabled Scientific Data Repository
There is a clear disconnect between chemical databases, publishers' data repositories, and semantic web knowledge bases. OSDR provides a basic chemistry data processing pipeline, including validation and standardization of chemical representation, but unlike PubChem, ChemSpider [26, 27], and others, it extends the list of supported types to reactions, crystals, and analytical data. Because of the ability to read and interpret chemical formats, OSDR provides chemical-indexing capabilities (the content can be searched by chemical structure, reaction, etc.) on top of regular searches by various alphanumeric properties. OSDR allows real-time data curation and will support ontology-based property assignment with subsequent complex searches. OSDR's deposition pipeline includes the data mining stage which, for example, allows text mining and chemical names to chemical structures conversion on the fly when a new document is deposited. The OSDR security model supports private, shared, and public data. Statistical models are usually built on versatile data collected from various data sources. It is widely known that the quality and domain of applicability of models is defined by those of the training datasets (the primary sources of such data are, e.g., PubChem and ChEMBL). OSDR, by incorporating a data mining and curation pipeline on top of integration with multiple data sources, provides a platform for the rapid composition of training datasets for immediate modeling. It was shown in multiple publications [28] that the quality of quantitative structure–activity relationship (QSAR) predictions highly depends on how the data used for model training was prepared as well as the type of descriptors used in model training and does not depend that much on the machine learning method.
13.3.2 Chemical Validation and Standardization Platform
The Chemical Validation and Standardization Platform (CVSP) [27] is an open source application developed for OpenPHACTS for chemical data validation and standardization. The original rules set was taken from the FDA Substances Registry System user manual [29] and extended with IUPAC rules. CVSP is already a part of the OSDR data processing pipeline and will be extended further, beyond chemical structures, into other types of data.
13.3.3 Format Adapters
While recent standardization attempts will eventually result in a set of adopted formats for data representation, historically (and especially before the appearance of structured and schema-driven formats such as XML and JSON), a number of various data formats were developed and adopted. Notable efforts include the development of SPL (stands for structured product labeling and is an HL7 standard for medical information exchange and a future format for application submission into United State Food and Drug Administration's Substance Registry System), ISA-tab and recently JSA-JSON, National Center for Biotechnology Information ASN.1, and others. On the other hand, while publishers mainly operate and accept ChemDraw CDX files with publications, major public databases such as PubChem and ChemSpider accept MOL and SDF formats, whereas for those involved with chemistry-related data science, the most convenient format is SMILES, InChI, InChIKey, or a set of internal or external identifiers. Some of these identifiers (e.g., InChIKey) are not reciprocally convertible, hence require existence of specific registries (e.g., InChI Resolver). Proprietary and in-house developed identifiers impose a similar challenge, but with an even more complicated situation as custom identifiers are never supported by publicly available registries. This quickly leads to the situation of creating multiple discrepancies on the level of data reformatting and exchange requiring a major effort to prepare data for deposition into PubChem or other databases. OSDR already supports various formats and is able to convert between them seamlessly on the fly at the time of import or export. The situation will only become more complicated when the FDA will start accepting applications for broad classes of chemicals in SPL format. With the experience gained by us in developing SPL representation of various classes of chemicals (substances, mixtures, proteins, structurally diverse substances, polymers, DNAs, RNAs, and various conjugates), OSDR already provides the ability to convert popular structure representations into SPL. Another problem is that most chemical data exist as CSV files with chemical structures encoded as CAS numbers, chemical names, SMILES, or InChIs and InChIKeys (in some cases). Chemical modeling software as well as other applications expect chemical structure to be represented as a connection graph which, for example should be able to extract/calculate fingerprints. Such a conversion may be trivial for SMILES and InChIs, but is problematic for chemical names, CAS and other registry numbers, InChIKeys, and other types of chemical identifiers. OSDR has the built in capability to load CSV files, specify column mapping to semantic type, and run real-time conversion with results being visually controlled. Once information is imported into a system, other operations such as export in various formats and modeling become possible. Another case publishing use is again connected to the fact that majority of electronic supplementary information exists as ChemDraw CDX files. There are multiple issues concerned with CDX files, including those with poor quality of drawing and misrepresentation of chemicals and reactions. OSDR reads and converts CDX files on the fly at deposition time and is able to flag erroneous chemistry using the built-in CVSP [27]. OSDR also allows export of all such imported/converted data in the SDF format, which is accepted by most public databases. Such functions can facilitate data depositions into those public databases which otherwise would require small businesses to buy costly software and resort to manual data preparation and curation, which is prohibitively expensive.
13.3.4 Open Platform for Data Acquisition, Curation, and Dissemination
Data are essential in science and data repositories well-supported by National Science Foundation (NSF) and other grants have been around for decades; yet they consist of merely file stores. The missing traits are (i) format awareness (the ability to visualize, index, and convert numerous domain-specific formats), (ii) secure data sharing, (iii) an ontological layer, (iv) real-time data quality checks (e.g., structure validation using CVSP), (v) integration with social networks (share, reuse, and build-on existing public profiles, including ORCID), and (vi) browser integration for ease of use and data submission. OSDR was designed with all these missing traits in mind and with proper implementation, it can create an ecosystem for the data market. This is a concept that rests on the assumption that should any goods (including information) become a commodity and proper tools operating on those commodities become available, then there are ample opportunities for new businesses to thrive on such a commodities market supplying various services around the supply–demand pipeline. For the data market, OSDR could help to provide chemicals for synthesis, chemical waste/by-product utilization, experimental data measurements, algorithm/model development (e.g., toxicology models for environmental or consumer product uses), and many others applications.
13.3.5 Dataledger
We will implement DataLedger, which represents an open source framework based on the approach and technology used in Blockchain [30] and Hyperledger [31], which will allow creating a secure, open, and distributed chain of data, metadata, and provenance events associated with information stored in OSDR. Such technology allows tracking and deredundifying data, operations on data, and authentic results, which will help to eliminate data plagiarism, a tremendous challenge for scientific research. By various estimates only 3–5% of collected experimental data ever find their way into publication and supplementary information, while the rest gradually decay in private collections or institutional data vaults. The methodology of proper research data sharing was developed as a part of FORCE11 effort, but technical implementation based on FAIR data principles [20] is lagging behind. We believe that the concept of an OSDR supported by Dataledger technology will greatly facilitate creation of the Science Data Market. The latter is based on the assumption that should any goods including intellectual goods become a commodity and if an unobstructed flow of such commodities is facilitated by respective systems and tools then a new opportunity for free trade becomes possible. There is clearly no such case for scientific data which are produced in great volumes and normally funded by taxpayers money and at best find their way into open access or closed access publications with no way to reproduce the original results. Yet liberating such scientific data would have a tremendous impact on scientific progress in general. If the process of data creation (e.g., experimental measurement or data mining) becomes fully attributable to those who conduct it and if the system facilitates proper data licencing and attribution at all subsequent stages, then scientific data become commodities and those who participate in every stage of this process should be allowed to at least gain an attribution or at most be rewarded intellectually and financially. We think that this could be incredibly valuable in making data relevant to toxicology available in this way.
13.4 Technical Details
OSDR represents a knowledge base for real-time management of various scientific data. It recognizes various formats including the most popular cheminformatics formats (MOL and SDF [32] for representing chemical substances, RXN and RDF [33] for representing reactions and metabolic pathways, CIF [34] for representing crystallographic data, PDB [35] for representing proteins, InChI [36], SMILES [37], CML [38] for representing various types of chemical information), office formats (Microsoft Word, Excel and PowerPoint), PDF, analytical data formats (JCAMP [39] and FID for spectral data), as well as general tabular data (CSV format, containing information about chemicals such as SMILES, InChI, or chemical name along with general identifiers, properties and other text and numerical data). The flexible and extensible architecture allows it to connect new plugins for handling additional data types.
The architecture of the system follows a regular pattern for deposition gateways into large chemical databases and is built on the assumption that raw data being deposited into a system have to undergo a series of parameterized transformations which in general are represented by a workflow consisting of various elements including blocks providing domain-specific functionality (e.g., format conversion, data import and export, automated chemical validation, and standardization [27]). The OSDR workflow supports extension of the core functionality with implementation of functional blocks which are connected into a system using.NET reflection [40] and MEF framework [41]. The architecture of the system is a result of our approximately 20 years of experience in building similar platforms for pharma companies and the broad scientific community (PubChem [42], ChemSpider [43], OpenPHACTS [13], National Chemistry Database Service [44]).
OSDR follows the Agile development approach that allows rapid development-deployment-customer feedback iterations. The system is being developed as a set of loosely coupled independent services. This approach, called microservices [45], Figure 13.5, as of today, is broadly accepted and considered to be suitable for development of systems with a wide range of complexity. The microservices approach allows adding new functionality to the existing system without affecting the rest of the system's stability. New modules and services therefore can be developed and added to the system by either the developers or the open source software/scientific community as the need for such arises. Another important consideration of this architectural approach is the ability to build technically heterogeneous systems that can use any programming language or framework given the precondition that they follow the industry-standard RESTful application communication conventions. RESTful services are build using the regular HTTP protocol that is universally supported and does not need additional hardware or any additional software libraries to support [46]. This feature allows disparate teams to build extension services using the programming expertise they have with the tools they are most comfortable with.

Figure 13.5 OSDR microservice overview.
OSDR is also built with a security-first principle in mind. While the whole system comprises a set of micro-services and APIs, access to everything is controlled on the basis of cryptographically strong and secure network protocols. They support not just control for the data access but also sharing of data between collaborating parties.
Integration with various cloud drives (currently GoogleDrive, DropBox, and Box) allows bidirectional secure exchange with information stored elsewhere and seamless import and export in various formats. An intuitive user interface provides ample capabilities for organizing the data in a way that corresponds to the organizational structure, field of research, ease of representation, and perception. The latest web frameworks make the system extremely lightweight, scalable, and able to run on all devices (desktops, tablets, handheld devices). The information presentation can be adjusted for the field of study, particular projects, and collaboration. Documents are represented with thumbnails with associated snippets of data and support full-screen previews.
The document mode (Figure 13.3) supports various property views and allows the entering of additional arbitrary or templated information. The built-in preview mode (Figure 13.4) allows reading of rich text documents directly into OSDR. Large sets of supported formats allow OSDR to store various data files that are usually used and produced in research next to each other. With the integrated format conversion ability, one needs to pay very little attention to a particular storage format, focusing instead on the information nature, provenance, and analysis. OSDR supports import from various external systems and websites including ChemSpider, Wikipedia, and arbitrary web pages with subsequent text and data mining. Complementary chemical conversion restores concise hierarchy representing complex areas of study – chemicals, syntheses, materials, and so on. Records and file information are organized into a set of info-boxes with simple and complex fields. Complex fields allow OSDR to store, visualize, and index virtually any datatypes which can be hierarchically organized. The ability to associate licenses allows it to “mix and match” both open, embargoed, and closed data to produce derivatives and share derived knowledge, still keeping track of all operations involved in data manipulations. The ability to assign keywords based on controlled vocabularies allows OSDR to navigate data in various alternative ways. Custom and standard property editors and visualizers then allow creation and representation of complex knowledge in a clear and concise manner.

Figure 13.6 Logical architecture of OSDR with cheminformatics and machine learning modules.
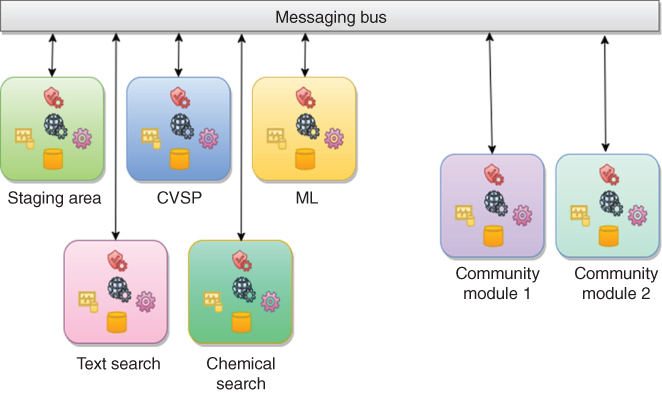
Figure 13.7 OSDR microservice-oriented architecture.
13.5 Future Work
The work on OSDR was started in November 2015 with an early prototype created in the summer of 2016. It was then realized that to keep the platform capable of responding to a modern informatics world challenges, the architecture of the system had to be completely changed and this has taken almost another year to implement. OSDR therefore currently consists of several major subsystems and a number of connected services (Figure 13.6). A staging area is a subsystem that supports real-time data management operations. CVSP (described earlier) is one of the microservices. Other services perform data import, export, conversion, processing, curation, and other functions. Information entry and editing is done in the user interface and conveyed to the main part of the system only through the RESTful Web API. Security control is currently implemented on top of IdentityServer. All system components are decoupled and interact either through RESTful API or using a system bus. Angular2 is used as a client framework, NET platform (currently a mix of .NET Framework 4.6.2 and .NET Core) is used for the server-side and MongoDB is used as the main database engine, although some services have their own specialized backup stores (e.g., Redis and ElasticSearch) (Figure 13.7). Microservice architecture is commonly associated with containerization technologies such as Docker [47] and Windows Containers [48] (Figure 13.8).
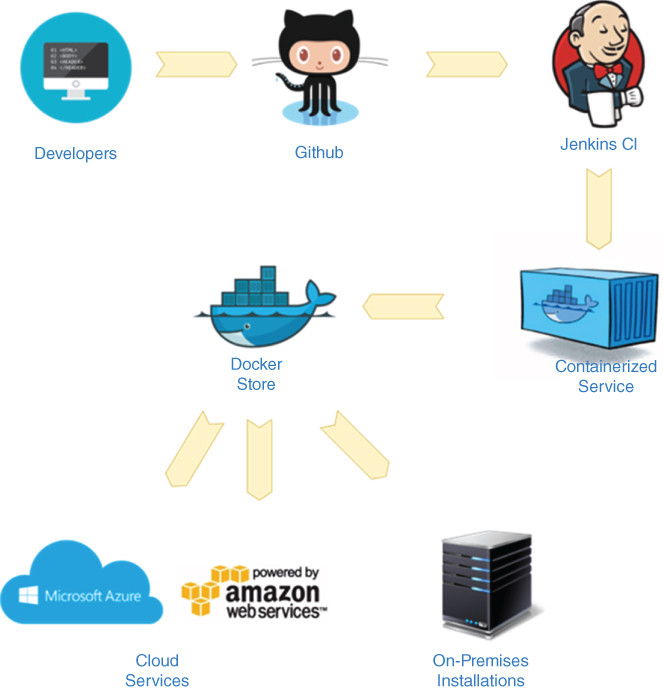
Figure 13.8 OSDR development workflow.
Each OSDR module, or service, is packaged as a container image that requires minimum configuration when deployed. The containers can be deployed using on-premises datacenters, on individual computers, or in a cloud services such AWS, Azure, or Google, using the same images that can published in the Docker Store repository, that similarly to Github, allows version control and convenient storage of the container images. Since OSDR, as a platform, consists of self-contained containerized services, the performance of the overall system can be adjusted by simply adding more container instances of the services that impede the whole system's performance. The containerized services can be dynamically spun up during peak hours or brought down to save costs by container orchestration mechanisms such as Docker Swarm [49] or Google Kubernetes [50]. These mechanisms observe the system's usage, or load, and automatically add or remove service instances, allowing for system elasticity. This way, any scale desired for the current installation and/or current scientific research process can be achieved.
OSDR is built using the modern CI/CD (continuous integration/continuous delivery) development paradigm to significantly shorten the software delivery time while ensuring the quality of the system. This is achieved through a succession of steps. Static code analysis tools are used at the development time to aid with the code checks while it is being written. Requiring that most, if not all, of the source code is covered by unit tests helps to ensure that the building blocks of the system are designed properly and function properly without hitches. The unit tests are run by a continuous integration server, such as Jenkins, at the time of each code check-in, so that the developer breaking any module would be notified immediately of any broken tests. A set of BDD (behavior-driven development) tests is being developed by the quality assurance team. The BDD tests describe the specific end-user scenarios. The tests can be written in a language closely resembling human-readable use cases. These tests are run automatically as well as part of the CI/CD pipeline with detailed reports indicating any potential problems.
13.5.1 Implementation of Ontology-Based Properties
The properties editor in OSDR allows in-place operations on associated data types and values, including hierarchical ones which are used not only to capture a value of the property but also associated metadata, for example, provenance data, associated conditions, and references which is essential for data quality assessment and control. Such property values are stored in JSON and are imported from external sources generated during processing or created as a part of the curation process. Almost the same JSON representation is being synchronized with ElasticSearch that provides an out-of-the-box rich and fast full text and property search experience. To validate and enforce value correctness, we will allow it to connect externally controlled vocabularies whose content is usually available as plain text, XML, or JSON dumps. We will also provide an editor for controlled vocabularies as a part of OSDR. The OSDR properties data model allows it to associate property type with its semantic value and by associating semantic type with one or more controlled vocabularies, we will enforce data validation. To improve user experience and make the curation process more powerful we will develop specialized editors for complex properties. For example, chemical structure, which is usually stored as SMILES or MOL, will be conveniently edited in the chemical sketcher. We also provide a way to organize property groups into “info-boxes” based on their semantic meaning. The info-box will have a “schema” (JSON schema) associated with it and we already have code under development which allows generating view/edit forms based on the JSON schema.
13.5.2 Implementation of an Advanced Search System
Currently, OSDR allows data acquisition from numerous sources and formats, including many domain-specific formats and office documents, and even capturing data and metadata from web pages. But as the number of supported formats, their complexity, and the size of the data in OSDR grows it becomes essential to provide an alternative navigation system based on domain-specific information (such as chemical substructure, similarity, clustering), associated keywords, powerful free-text query variants, ontological terms, complex property values, and so on. We will implement an advanced search system for all information in OSDR and provide alternative navigation capabilities. Current searching in OSDR is based on ElasticSearch which is being populated with a replica of JSON representing respective objects in the staging area. We will expand search capabilities in the following ways. First, we will generate search forms based on the JSON schema associated with various info-boxes thus allowing one to use particular search criteria based on the area of research interest. Secondly, we will provide a set of “orthogonal” searches to slice data collections based on field types associated with particular data types. Combination of such searches with previous full-text searches will allow navigation of all data not just in hierarchical manner (which is connected to how the data are arranged in a system) but create data slices based on specific criteria. Third, we will implement chemical structure and reaction searches using third party open source software (Bingo from EPAM).
13.5.3 Implementation of a Scientist Profile, Advanced Security, Data Sharing Capabilities and Notifications Framework
We will extend basic authentication and integration with social networks by developing an OSDR Scientist Profile which extends the one used by ORCID [51] and other publishing user profiles. To facilitate collaboration between scientists as well as their groups using data stored in OSDR, we will complete implementation of the data sharing and security layers which will fully support fine-grained access to the data on all levels. We will let scientists invite and designate collaborators, expose data and send new data sharing requests, provide data quality ranking and usage statistics, and provide a comprehensive notification framework. To achieve this, we will start by extending the OSDR user profile with information specific to OSDR. We will then add a configuration layer on top of the OSDR basic schema to allow specification of access rules which can be enforced at organization and user levels. When respective access to such particular functions or area of OSDR is requested, we will use OAuth protocol to solicit such access from the resource owner. We will also implement a simple notification framework to notify resource owners of concerned actions of other system users.
Further, we will work on integration with various social networks, cloud storage systems, and existing data repositories. We plan to expand the properties model by choosing and adapting taxonomies and ontologies relevant for biomedical information for example (BAO [52, 53]). We will also implement a full secure data sharing and collaboration mechanism that is based on the public key infrastructure [54], OpenID, and OAuth2. As we expect OSDR to grow and start gathering more information, the scientific data in such a system will become a commodity and as such will create an opportunity to further develop a “data market.”
OSDR presents an opportunity for structure-activity datasets relating to toxicology to be accumulated. In turn, the standardization will provide datasets already in a format for descriptor generation and enable machine learning model construction. Such models could hence be stored in OSDR and potentially shared with collaborators. The benefits of using such open data repositories for academics could be that it will facilitate data sharing in a cost-effective manner. For small companies, it will provide industry-quality technology that is readily accessible. If we are to leverage the growth in toxicology-related datasets in recent years, we need them to be accessed for machine learning. OSDR will provide this capability.
References
- 1 Ekins, S., Hohman, M., and Bunin, B.A. (2011) Pioneering use of the cloud for development of the collaborative drug discovery (CDD) database, in Collaborative Computational Technologies for Biomedical Research (eds S. Ekins, M.A.Z. Hupcey, and A.J. Williams), John Wiley & Sons, Inc., Hoboken, NJ.
- 2 Hohman, M., Gregory, K., Chibale, K. et al. (2009) Novel web-based tools combining chemistry informatics, biology and social networks for drug discovery. Drug Discov. Today, 14, 261–270.
- 3 Bost, F., Jacobs, R.T., and Kowalczyk, P. (2010) Informatics for neglected diseases collaborations. Curr. Opin. Drug Discov. Dev., 13, 286–296.
- 4 Cressey, D. (2011) Traditional drug-discovery model ripe for reform. Nature, 471, 17–18.
- 5 Guiguemde, W.A., Shelat, A.A., Bouck, D. et al. (2010) Chemical genetics of Plasmodium falciparum. Nature, 465, 311–315.
- 6 Gamo, F.-J., Sanz, L.M., Vidal, J. et al. (2010) Thousands of chemical starting points for antimalarial lead identification. Nature, 465, 305–310.
- 7 Fidock, D.A. (2010) Drug discovery: priming the antimalarial pipeline. Nature, 465, 297–298.
- 8 Ekins, S. and Williams, A.J. (2010) Meta-analysis of molecular property patterns and filtering of public datasets of antimalarial “hits” and drugs. MedChemCommun, 1, 325–330.
- 9 Drewry, D.H., Willson, T.M., and Zuercher, W.J. (2014) Seeding collaborations to advance kinase science with the GSK published kinase inhibitor set (PKIS). Curr. Top. Med. Chem., 14, 340–342.
- 10 Bento, A.P., Gaulton, A., Hersey, A. et al. (2014) The ChEMBL bioactivity database: an update. Nucleic Acids Res., 42, D1083–D1090.
- 11 Bradley, J.-C., Guha, R., Lang, A. et al. (2009) Beautifying data in the real world, in Beautiful data (eds T. Segaran and J. Hammerbacher), O'Reilly Media Inc., Sebastopol, CA.
- 12 Bradley, J.C., Lancashire, R.J., Lang, A.S., and Williams, A.J. (2009) The spectral game: leveraging open data and crowdsourcing for education. J. Cheminform., 1, 9.
- 13 http://www.openphacts.org/. OpenPHACTS, http://www.openphacts.org/ (accessed August 14, 2017).
- 14 Azzaoui, K., Jacoby, E., Senger, S. et al. (2013) Scientific competency questions as the basis for semantically enriched open pharmacological space development. Drug Discov. Today, 18 (17–18), 843–852.
- 15 Hunter, J. (2011) Precompetitive collaboration in the pharmaceutical industry, in Collaborative computational Technologies for Biomedical Research (eds S. Ekins, M.A.Z. Hupcey, and A.J. Williams), John Wiley & Sons, Inc., Hoboken, NJ, pp. 55–84.
- 16 Hunter, J. and Stephens, S. (2010) Is open innovation the way forward for big pharma? Nat. Rev. Drug Discov., 9, 87–88.
- 17 Hunter, A.J. (2008) The innovative medicines initiative: a pre-competitive initiative to enhance the biomedical science base of Europe to expedite the development of new medicines for patients. Drug Discov. Today, 13, 371–373.
- 18 ChEMBL, http://www.ebi.ac.uk/chembldb/index.php (accessed August 14, 2017).
- 19 Huang, R., Xia, M., Sakamuru, S. et al. (2016) Modelling the Tox21 10 K chemical profiles for in vivo toxicity prediction and mechanism characterization. Nat. Commun., 7, 10425.
- 20 Anon, The fair data principles, https://www.force11.org/group/fairgroup/fairprinciples (accessed August 14, 2017).
- 21 Kim, S., Thiessen, P.A., Bolton, E.E. et al. (2016) PubChem substance and compound databases. Nucleic Acids Res., 44, D1202–D1213.
- 22 Degtyarenko, K., de Matos, P., Ennis, M. et al. (2008) ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res., 36, D344–D350.
- 23 Anon, Figshare, http://figshare.com/ (accessed August 14, 2017).
- 24 Anon (2017) Mendeley, https://data.mendeley.com/ (accessed August 14, 2017).
- 25 Tkachenko, V. (2017) OSDR, https://github.com/scidatasoft/OSDR (accessed August 14, 2017).
- 26 Pence, H.E. and Williams, A.J. (2010) ChemSpider: an online chemical information resource. J. Chem. Educ., 87, 1123–1124.
- 27 Karapetyan, K., Batchelor, C., Sharpe, D. et al. (2015) The chemical validation and standardization platform (CVSP): large-scale automated validation of chemical structure datasets. J. Cheminform., 7, 30.
- 28 Fourches, D., Muratov, E., and Tropsha, A. (2010) Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research. J. Chem. Inf. Model., 50, 1189–1204.
- 29 FDA (2007) Food and Drug Administration Substance Registration System Standard Operating Procedure, https://www.fda.gov/downloads/ForIndustry/DataStandards/SubstanceRegistrationSystem-UniqueIngredientIdentifierUNII/ucm127743.pdf (accessed August 14, 2017).
- 30 Anon, Blockchain, https://en.wikipedia.org/wiki/Blockchain_(database) (accessed August 14, 2017).
- 31 Anon, Hyperledger, https://www.hyperledger.org/ (accessed August 14, 2017).
- 32 Anon, Chemical table file, https://en.wikipedia.org/wiki/Chemical_table_file (accessed August 14, 2017).
- 33 Anon, MDL MOLfiles, RGfiles, SDfiles, Rxnfiles, RDfiles formats. https://docs.chemaxon.com/display/docs/MDL+MOLfiles,+RGfiles,+SDfiles,+Rxnfiles,+RDfiles+formats (accessed August 14, 2017).
- 34 Anon, Common intermediate format, https://en.wikipedia.org/wiki/Common_Intermediate_Format (accessed August 14, 2017).
- 35 Anon, File format, http://www.wwpdb.org/documentation/file-format (accessed August 14, 2017).
- 36 Anon, Inchi, https://iupac.org/who-we-are/divisions/division-details/inchi/ (accessed August 14, 2017).
- 37 Anon, Simplified molecular-input line-entry system, https://en.wikipedia.org/wiki/Simplified_molecular-input_line-entry_system (accessed August 14, 2017).
- 38 Anon, Chemical markup language, https://en.wikipedia.org/wiki/Chemical_Markup_Language (accessed August 14, 2017).
- 39 Anon, JCAMP, http://www.jcamp-dx.org/ (accessed August 14, 2017).
- 40 Anon, Reflection in the.NET framework, https://msdn.microsoft.com/en-us/library/f7ykdhsy%28v=vs.110%29.aspx?f=255&MSPPError=-2147217396 (accessed August 14, 2017).
- 41 Anon, Managed extensibility framework (MEF), https://msdn.microsoft.com/en-us/library/dd460648(v=vs.110).aspx (accessed August 14, 2017).
- 42 Anon, The PubChem Database, http://pubchem.ncbi.nlm.nih.gov/ (accessed August 14, 2017).
- 43 ChemSpider, http://www.chemspider.com (accessed August 14, 2017).
- 44 Anon, National Chemistry Database Service, http://cds.rsc.org/ (accessed August 14, 2017).
- 45 Fowler M. (2014) Microservices, https://martinfowler.com/articles/microservices.html (accessed August 14, 2017).
- 46 Anon, RESTful services, https://en.wikipedia.org/wiki/Representational_state_transfer (accessed August 14, 2017).
- 47 Anon, Docker containers, https://www.docker.com/what-docker (accessed August 14, 2017).
- 48 Microsoft Windows Containers (2016) https://docs.microsoft.com/en-us/virtualization/windowscontainers/about/ (accessed August 14, 2017).
- 49 Anon, Docker Swarm, https://www.docker.com/products/docker-swarm (accessed August 14, 2017).
- 50 Anon, Google Kubernetes, https://kubernetes.io/ (accessed August 14, 2017).
- 51 Anon (2017), ORCID, https://orcid.org/ (accessed August 14, 2017).
- 52 BAO, http://bioassayontology.org (accessed August 14, 2017).
- 53 Visser, U., Abeyruwan, S., Vempati, U. et al. (2011) BioAssay Ontology (BAO): a semantic description of bioassays and high-throughput screening results. BMC Bioinformatics, 12, 257.
- 54 Anon, Public key infrastructure. PKI, https://en.wikipedia.org/wiki/Public_key_infrastructure (accessed August 14, 2017).
