In the previous section, we saw that the basic operation of the variable elimination algorithm is the manipulation of the factors. First, we create a factor ![]() by multiplying existing factors. Then, we eliminate a variable in
by multiplying existing factors. Then, we eliminate a variable in ![]() to generate a new factor
to generate a new factor ![]() , which is then used to create another factor. From a different perspective, we can say that a factor
, which is then used to create another factor. From a different perspective, we can say that a factor ![]() is a data structure, which takes messages
is a data structure, which takes messages ![]() generated by the other factor
generated by the other factor ![]() , and generates a message
, and generates a message ![]() which is used by the other factor
which is used by the other factor ![]() .
.
Before we go into a detailed discussion of the belief propagation algorithm, let's discuss the graphical model that provides the basic framework for it, the clique tree, also known as the junction tree.
The clique tree (![]() ) is an undirected graph over a set of factors
) is an undirected graph over a set of factors ![]() , where each node represents a cluster of random variables and the edges connect the clusters, whose scope has a nonempty intersection. Thus, each edge between a pair of clusters
, where each node represents a cluster of random variables and the edges connect the clusters, whose scope has a nonempty intersection. Thus, each edge between a pair of clusters ![]() and
and ![]() is associated with a sepset
is associated with a sepset ![]() . For each cluster
. For each cluster ![]() , we also define the cluster potential
, we also define the cluster potential ![]() , which is the factor representing all the variables present in it.
, which is the factor representing all the variables present in it.
This can be generalized. Let's assume there is a variable X, such that ![]() and
and ![]() . Then, X is also present in every cluster in the path between
. Then, X is also present in every cluster in the path between ![]() and
and ![]() in
in ![]() . This is known as the running intersection property. We can see an example in the following figure:
. This is known as the running intersection property. We can see an example in the following figure:

Fig 3.4: A simple cluster tree with clusters ![]() and
and ![]() . The sepset associated with the edge is
. The sepset associated with the edge is ![]() .
.
In pgmpy, we can define a clique tree or junction tree in the following way:
# Firstly import JunctionTree class
In [1]: from pgmpy.models import JunctionTree
In [2]: junction_tree = JunctionTree()
# Each node in the junction tree is a cluster of random variables
# represented as a tuple
In [3]: junction_tree.add_nodes_from([('A', 'B', 'C'),
('C', 'D')])
In [4]: junction_tree.add_edge(('A', 'B', 'C'), ('C', 'D'))
In [5]: junction_tree.add_edge(('A', 'B', 'C'), ('D', 'E', 'F'))
ValueError: No sepset found between these two edges.In the previous section on variable elimination, we saw that an induced graph created by variable elimination is a chordal graph. The converse of it also holds true; that is, any chordal graph can be used as a basis for inference.
We previously discussed chordal graphs, triangulation techniques (the process of constructing a chordal graph that incorporates an existing graph), and their implementation in pgmpy. To construct a clique tree from the chordal graph, we need to find the maximal cliques in it. There are multiple ways of doing this. One of the simplest methods is the maximum cardinality search (which we discussed in the previous section) to obtain maximal cliques.
Then, these maximal cliques are assigned as nodes in the clique tree. Finally, to determine the edges of the clique tree, we use the maximum spanning tree algorithm. We build an undirected graph whose nodes are maximal cliques in ![]() , where every pair of nodes
, where every pair of nodes ![]() ,
, ![]() is connected by an edge whose weight is
is connected by an edge whose weight is ![]() . Then, by applying the maximum spanning tree algorithm, we find a tree in which the weight of edges is at maximum.
. Then, by applying the maximum spanning tree algorithm, we find a tree in which the weight of edges is at maximum.
The cluster potential for each cluster in the clique tree can be computed as the product of the factors associated with each node of the cluster. For example, in the following figure, ![]() (the cluster potential associated with cluster (A, B, C) is computed as the product of P(A), P(C), and
(the cluster potential associated with cluster (A, B, C) is computed as the product of P(A), P(C), and ![]() . To compute
. To compute ![]() (the cluster potential associated with (B, D, E), we use
(the cluster potential associated with (B, D, E), we use ![]() , P(B), and P(D). P(B) is computed by marginalizing
, P(B), and P(D). P(B) is computed by marginalizing ![]() with respect to A and C.
with respect to A and C.

Fig 3.5: The cluster potential of the clusters present in the clique tree
These steps can be summarized as follows:
-
Triangulate the graph G over factor
 to create a chordal graph
to create a chordal graph  .
.
-
Find the maximal cliques in
 and assign them as nodes to an undirected graph.
and assign them as nodes to an undirected graph.
- Assign weights to the edges between two nodes of the undirected graph as the numbers of elements in the sepset of the two nodes.
- Construct the clique tree using the maximum spanning tree algorithm.
- Compute the cluster potential for each cluster as the product of factors associated with the nodes present in the cluster.
In pgmpy, each graphical model class has a method called to_junction_tree(), which creates a clique tree (or junction tree) corresponding to the graphical model. Here's an example:
In [1]: from pgmpy.models import BayesianModel, MarkovModel
In [2]: from pgmpy.factors import TabularCPD, Factor
# Create a bayesian model as we did in the previous chapters
In [3]: model = BayesianModel(
[('rain', 'traffic_jam'),
('accident', 'traffic_jam'),
('traffic_jam', 'long_queues'),
('traffic_jam', 'late_for_school'),
('getting_up_late', 'late_for_school')])
In [4]: cpd_rain = TabularCPD('rain', 2, [[0.4], [0.6]])
In [5]: cpd_accident = TabularCPD('accident', 2, [[0.2], [0.8]])
In [6]: cpd_traffic_jam = TabularCPD(
'traffic_jam', 2,
[[0.9, 0.6, 0.7, 0.1],
[0.1, 0.4, 0.3, 0.9]],
evidence=['rain', 'accident'],
evidence_card=[2, 2])
In [7]: cpd_getting_up_late = TabularCPD('getting_up_late', 2,
[[0.6], [0.4]])
In [8]: cpd_late_for_school = TabularCPD(
'late_for_school', 2,
[[0.9, 0.45, 0.8, 0.1],
[0.1, 0.55, 0.2, 0.9]],
evidence=['getting_up_late',
'traffic_jam'],
evidence_card=[2, 2])
In [9]: cpd_long_queues = TabularCPD('long_queues', 2,
[[0.9, 0.2],
[0.1, 0.8]],
evidence=['traffic_jam'],
evidence_card=[2])
In [10]: model.add_cpds(cpd_rain, cpd_accident,
cpd_traffic_jam, cpd_getting_up_late,
cpd_late_for_school, cpd_long_queues)
In [11]: junction_tree_bm = model.to_junction_tree()
In [12]: type(junction_tree_bm)
Out[12]: pgmpy.models.JunctionTree.JunctionTree
In [13]: junction_tree_bm.nodes()
Out[13]:
[('traffic_jam', 'getting_up_late', 'late_for_school'),
('traffic_jam', 'rain', 'accident'),
('traffic_jam', 'long_queues')]
In [14]: junction_tree_bm.edges()
Out[14]:
[(('traffic_jam', 'long_queues'),
('traffic_jam', 'late_for_school', 'getting_up_late')),
(('traffic_jam', 'long_queues'), ('traffic_jam', 'rain',
'accident'))]Let's go back to the previous example of the Bayesian network for the late- for school- example:

Fig 3.6: Bayesian network for a student being late for school.
In the previous section, we saw how to construct a clique tree for this Bayesian network. The following figure shows the clique tree for this network:

Fig 3.7: Clique tree constructed from the Bayesian network presented in Fig 3.3
As we discussed earlier, in the belief propagation algorithm, we would be considering factor ![]() to be a computational data structure that would take a message
to be a computational data structure that would take a message ![]() generated from a factor
generated from a factor ![]() , and produce
, and produce ![]() , which can be further passed on to another factor
, which can be further passed on to another factor ![]() , and so on.
, and so on.
Let's go into the details of what each message term (![]() and
and ![]() ) means. Let's begin with a very simple example of finding the probability of being late for school (L). To compute the probability of L, we need to eliminate all the random variables, such as accident (A), rain (R), traffic jam (J), getting up late (G), and long queues (Q). We can see that variables A and R are present only in cluster
) means. Let's begin with a very simple example of finding the probability of being late for school (L). To compute the probability of L, we need to eliminate all the random variables, such as accident (A), rain (R), traffic jam (J), getting up late (G), and long queues (Q). We can see that variables A and R are present only in cluster ![]() and Q is present only in
and Q is present only in ![]() , but J is present in all three clusters, namely
, but J is present in all three clusters, namely ![]() ,
, ![]() , and
, and ![]() . So, both A and R can be eliminated from
. So, both A and R can be eliminated from ![]() by just marginalizing
by just marginalizing ![]() with respect to A and R. Similarly, we could eliminate Q from
with respect to A and R. Similarly, we could eliminate Q from ![]() . However, to eliminate J, we can't just eliminate it from
. However, to eliminate J, we can't just eliminate it from ![]() ,
, ![]() , or
, or ![]() alone. Instead, we need contributions from all three.
alone. Instead, we need contributions from all three.
Eliminating the random variables A and R by marginalizing the cluster potential ![]() corresponding to
corresponding to ![]() , we get the following:
, we get the following:
Similarly, marginalizing the cluster potential ![]() corresponding to
corresponding to ![]() with respect to Q, we get the following:
with respect to Q, we get the following:
Now, to eliminate J and G, we must use ![]() ,
, ![]() , and
, and ![]() . Eliminating J and G, we get the following:
. Eliminating J and G, we get the following:
From the perspective of message passing, we can see that ![]() produces an output message
produces an output message ![]() . Similarly,
. Similarly, ![]() produces a message
produces a message ![]() . These messages are then used as input messages for
. These messages are then used as input messages for ![]() to compute the belief for a cluster
to compute the belief for a cluster ![]() . Belief for a cluster
. Belief for a cluster ![]() is defined as the product of the cluster potential
is defined as the product of the cluster potential ![]() with all the incoming messages to that cluster. Thus:
with all the incoming messages to that cluster. Thus:
So, we can re-frame the following equation:
It can be re-framed as follows:
Fig 3.8 shows message propagation from clusters ![]() and
and ![]() to cluster
to cluster ![]() :
:
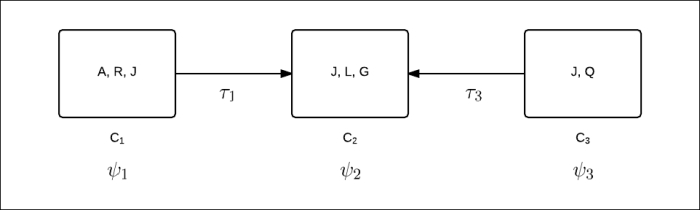
Fig 3.8: Message propagation from clusters ![]() and
and ![]() to cluster
to cluster ![]() :
:
Now, let's consider another example, where we compute the probability of long queues (Q). We have to eliminate all the other random variables, except Q. Using the same approach as discussed earlier, first marginalize ![]() with respect to A and R, and compute
with respect to A and R, and compute ![]() as follows:
as follows:
As discussed earlier, to eliminate the variable J, we need contributions from ![]() ,
, ![]() , and
, and ![]() , so we can't simply eliminate J from
, so we can't simply eliminate J from ![]() . The other two random variables L and G are only present in
. The other two random variables L and G are only present in ![]() , so we can easily eliminate them from
, so we can easily eliminate them from ![]() . However, to eliminate L and G from
. However, to eliminate L and G from ![]() , we can't simply marginalize
, we can't simply marginalize ![]() . We have to consider the contribution of
. We have to consider the contribution of ![]() (the outgoing message from
(the outgoing message from ![]() ) as well, because J was present in both the clusters
) as well, because J was present in both the clusters ![]() and
and ![]() . Thus, eliminating L and G would create
. Thus, eliminating L and G would create ![]() as follows:
as follows:
Finally, we can eliminate J by marginalizing the following belief ![]() of
of ![]() :
:
We can eliminate it as follows:
Fig 3.9 shows a message passing from ![]() to
to ![]() and
and ![]() to
to ![]() :
:

Fig 3.9: Message passing from ![]() to
to ![]() and
and ![]() to
to ![]()
In the previous examples, we saw how to perform variable elimination in a clique tree. This algorithm can be stated in a more generalized form. We saw that variable elimination in a clique tree induces a directed flow of messages between the clusters present in it, with all the messages directed towards a single cluster, where the final computation is to be done. This final cluster can be considered as the root. In our first example, the root was ![]() , while in the second example, it was
, while in the second example, it was ![]() . The notions of directionality and root also create the notions of upstream and downstream. If
. The notions of directionality and root also create the notions of upstream and downstream. If ![]() is on the path from
is on the path from ![]() to the root, then we can say that
to the root, then we can say that ![]() is upstream from
is upstream from ![]() , and
, and ![]() is downstream from
is downstream from ![]() :
:

Fig 3.10: ![]() is upstream from
is upstream from ![]() and
and ![]() is downstream from
is downstream from ![]()
We also saw in the second example that ![]() was not able to send messages to
was not able to send messages to ![]() until it received the message from
until it received the message from ![]() , as the generation of
, as the generation of ![]() also depends on
also depends on ![]() . This introduces the notion of a ready cluster. A cluster is said to be ready to transmit messages to its upstream cliques if it has received all the incoming messages from its downstream cliques.
. This introduces the notion of a ready cluster. A cluster is said to be ready to transmit messages to its upstream cliques if it has received all the incoming messages from its downstream cliques.
The message ![]() from the cluster j to the cluster i can be defined as the factor formed by the following sum product message passing computation:
from the cluster j to the cluster i can be defined as the factor formed by the following sum product message passing computation:

We can now define the terms belief ![]() of a cluster
of a cluster ![]() . It is defined as the product of all the incoming messages
. It is defined as the product of all the incoming messages ![]() from its neighbors with its own cluster potential:
from its neighbors with its own cluster potential:
Here, j is the upstream from i.
All these discussions for running the algorithm can be summarized in the following steps:
- Identify the root (this is the cluster where the final computation is to be made).
- Start with the leaf nodes of the tree. The output message of these nodes can be computed by marginalizing its belief. The belief for the leaf node would be its cluster potential as there would be no incoming message.
- As and when the other clusters of the clique tree become ready, compute the outgoing message and propagate them upstream.
- Repeat step 3 until the root node has received all the incoming messages.
In the previous section, we discussed how to compute the probability of any variable using belief propagation. Now, let's look at the broader picture. What if we wanted to compute the probability of more than one random variable? For example, say we want to know the probability of long queues as well as a traffic jam. One naive solution would be to do a belief propagation in the clique tree by considering each cluster as a root. However, can we do better?
Consider the previous two examples we have discussed. The first one had ![]() as the root, while the other had
as the root, while the other had ![]() . We saw that in both cases, message computed from the cluster
. We saw that in both cases, message computed from the cluster ![]() to the cluster
to the cluster ![]() (that is
(that is ![]() ) is the same, irrespective of the root node. Generalizing this, we can conclude that the message
) is the same, irrespective of the root node. Generalizing this, we can conclude that the message ![]() from the cluster
from the cluster ![]() to the cluster
to the cluster ![]() will be the same as long as the root is on the
will be the same as long as the root is on the ![]() side and vice versa. Thus, for a given edge in the clique tree between two clusters
side and vice versa. Thus, for a given edge in the clique tree between two clusters ![]() and
and ![]() , we have only two messages to compute, depending on the directionality of the edges (
, we have only two messages to compute, depending on the directionality of the edges (![]() and
and ![]() ). For a given clique tree with c clusters, we have
). For a given clique tree with c clusters, we have ![]() edges between these clusters. Thus, we only need to compute
edges between these clusters. Thus, we only need to compute ![]() messages.
messages.
As we have seen in the previous section, a cluster can propagate a message upstream as soon as it is ready, that is, when it has received all the incoming messages from downstream. So, we can compute both messages for each edge asynchronously. This can be done in two phases, one being an upward pass and the other being a downward pass. In the upward pass (Fig 3.11), we consider a cluster as a root and send all the messages to the root. Once the root has all the messages, we can compute its belief. For the downward pass (Fig 3.12), we can compute appropriate messages from the root to its children using its belief. This phase will continue until there is no message to be transmitted, that is, until we have reached the leaf nodes. This is shown in Fig 3.11:

Fig 3.11: Upward pass
Fig 3.11 shows an upward pass where cluster {E, F, G} is considered as the root node. All the messages from the other nodes are transmitted towards it.
The following figure shows a downward pass where the appropriate message from the root is transmitted to all the children. This will continue until all the leaves are reached:

Fig 3.12: Downward pass
When both, the upward pass and the downward pass are completed, all the adjacent clusters in the clique tree are said to be calibrated. Two adjacent clusters i and j are said to be calibrated when the following condition is satisfied:

In a broader sense, it can be said that the clique tree is calibrated. When a clique tree is calibrated, we have two types of beliefs, the first being cluster beliefs and the second being sepset beliefs. The sepset belief for a sepset ![]() can be defined as follows:
can be defined as follows:

Until now, we have viewed message passing in the clique tree from the perspective of variable elimination. In this section, we will see the implementation of message passing from a different perspective, that is, from the perspective of clique beliefs and sepset beliefs. Before we go into details of the algorithm, let's discuss another important operation on the factor called factor division.
A factor division between two factors ![]() and
and ![]() , where both X and Y are disjoint sets, is defined as follows:
, where both X and Y are disjoint sets, is defined as follows:

Here, we define ![]() . This operation is similar to the factor product, except that we divide instead of multiplying. Further, unlike the factor product, we can't divide factors not having any common variables in their scope. For example, consider the following two factors:
. This operation is similar to the factor product, except that we divide instead of multiplying. Further, unlike the factor product, we can't divide factors not having any common variables in their scope. For example, consider the following two factors:
|
a |
b |
|
|---|---|---|
|
|
|
0 |
|
|
|
1 |
|
|
|
2 |
|
|
|
3 |
|
|
|
4 |
|
|
|
5 |
|
b |
|
|---|---|
|
|
0 |
|
|
1 |
|
|
2 |
Dividing ![]() by
by ![]() , we get the following:
, we get the following:
|
a |
b |
|
|---|---|---|
|
|
|
0 |
|
|
|
1 |
|
|
|
1 |
|
|
|
0 |
|
|
|
4 |
|
|
|
2.5 |
In pgmpy, factor division can implemented as follows:
In [1]: from pgmpy.factors import Factor In [2]: phi1 = Factor(['a', 'b'], [2, 3], range(6)) In [3]: phi2 = Factor(['b'], [3], range(3)) In [4]: psi = phi1 / phi2 In [5]: print(psi) ╒═════╤═════╤════════════╕ │ a │ b │ phi(a,b) │ ╞═════╪═════╪════════════╡ │ a_0 │ b_0 │ 0.0000 │ ├─────┼─────┼────────────┤ │ a_0 │ b_1 │ 1.0000 │ ├─────┼─────┼────────────┤ │ a_0 │ b_2 │ 1.0000 │ ├─────┼─────┼────────────┤ │ a_1 │ b_0 │ 0.0000 │ ├─────┼─────┼────────────┤ │ a_1 │ b_1 │ 4.0000 │ ├─────┼─────┼────────────┤ │ a_1 │ b_2 │ 2.5000 │ ╘═════╧═════╧════════════╛
Let's go back to our original discussion regarding message passing using division. As we saw earlier, for any edge between clusters ![]() and
and ![]() , we need to compute two messages
, we need to compute two messages ![]() and
and ![]() . Let's assume that the first message was passed from
. Let's assume that the first message was passed from ![]() to
to ![]() , that is,
, that is, ![]() . So, a return message from
. So, a return message from ![]() to
to ![]() would only be passed when
would only be passed when ![]() has received all the messages from its neighbors.
has received all the messages from its neighbors.
Once ![]() has received all the messages from its neighbors, we can compute its belief
has received all the messages from its neighbors, we can compute its belief ![]() as follows:
as follows:
In the previous section, we also saw that the message from ![]() to
to ![]() can be computed as follows:
can be computed as follows:

From the preceding mathematical formulation, we can deduce that the belief of ![]() , that is,
, that is, ![]() , can't be used to compute the message from
, can't be used to compute the message from ![]() to
to ![]() as it would already include the message from
as it would already include the message from ![]() to
to ![]() in it:
in it:
That is:
Thus, from the preceding equation, we can conclude that the message from ![]() to
to ![]() can be computed by simply dividing the final belief of
can be computed by simply dividing the final belief of ![]() , that is,
, that is, ![]() , with the message from
, with the message from ![]() to
to ![]() , that is,
, that is, ![]() :
:

Finally, the message passing algorithm using this process can be summarized as follows:
-
For each cluster
 , initialize the initial cluster belief
, initialize the initial cluster belief  as its cluster potential
as its cluster potential  and sepset potential between adjacent clusters
and sepset potential between adjacent clusters  and
and  , that is,
, that is,  as 1.
as 1.
-
In each iteration, the cluster belief
 is updated by multiplying it with the message from its neighbors, and the sepset potential
is updated by multiplying it with the message from its neighbors, and the sepset potential  is used to store the previous message passed along the edge (
is used to store the previous message passed along the edge ( ), irrespective of the direction of the message.
), irrespective of the direction of the message.
- Whenever a new message is passed along an edge, it is divided by the old message to ensure that we don't count this message twice (as we discussed earlier).
Steps 2 and 3 can formally be stated in the following way for each iteration:

-
Here, we marginalize the belief to get the message passed. However, as we discussed earlier, this message will include a message from
 to
to  in it, so divide it by the previous message stored in
in it, so divide it by the previous message stored in  :
:

- Update the belief by multiplying it with the message from its neighbors:

- Update the sepset belief:

-
Repeat steps 2 and 3 until the tree is calibrated for each adjacent edge (
 ):
):

As this algorithm updates the belief of a cluster using the beliefs of its neighbors, we call it the belief update message passing algorithm. It is also known as the Lauritzen-Spiegelhalter algorithm.
In pgmpy, this can be implemented as follows:
In [1]: from pgmpy.models import BayesianModel
In [2]: from pgmpy.factors import TabularCPD, Factor
In [3]: from pgmpy.inference import BeliefPropagation
# Create a bayesian model as we did in the previous chapters
In [4]: model = BayesianModel(
[('rain', 'traffic_jam'),
('accident', 'traffic_jam'),
('traffic_jam', 'long_queues'),
('traffic_jam', 'late_for_school'),
('getting_up_late', 'late_for_school')])
In [5]: cpd_rain = TabularCPD('rain', 2, [[0.4], [0.6]])
In [6]: cpd_accident = TabularCPD('accident', 2, [[0.2], [0.8]])
In [7]: cpd_traffic_jam = TabularCPD('traffic_jam', 2,
[[0.9, 0.6, 0.7, 0.1],
[0.1, 0.4, 0.3, 0.9]],
evidence=['rain',
'accident'],
evidence_card=[2, 2])
In [8]: cpd_getting_up_late = TabularCPD('getting_up_late', 2,
[[0.6], [0.4]])
In [9]: cpd_late_for_school = TabularCPD(
'late_for_school', 2,
[[0.9, 0.45, 0.8, 0.1],
[0.1, 0.55, 0.2, 0.9]],
evidence=['getting_up_late','traffic_jam'],
evidence_card=[2, 2])
In [10]: cpd_long_queues = TabularCPD('long_queues', 2,
[[0.9, 0.2],
[0.1, 0.8]],
evidence=['traffic_jam'],
evidence_card=[2])
In [11]: model.add_cpds(cpd_rain, cpd_accident,
cpd_traffic_jam, cpd_getting_up_late,
cpd_late_for_school, cpd_long_queues)
In [12]: belief_propagation = BeliefPropagation(model)
# To calibrate the clique tree, use calibrate() method
In [13]: belief_propagation.calibrate()
# To get cluster (or clique) beliefs use the corresponding getters
In [14]: belief_propagation.get_clique_beliefs()
Out[14]:
{('traffic_jam', 'late_for_school', 'getting_up_late'): <Factor representing phi(getting_up_late:2, late_for_school:2, traffic_jam:2) at 0x7f565ee0db38>,
('traffic_jam', 'long_queues'): <Factor representing phi(long_queues:2, traffic_jam:2) at 0x7f565ee0dc88>,
('traffic_jam', 'rain', 'accident'): <Factor representing phi(rain:2, accident:2, traffic_jam:2) at 0x7f565ee0d4a8>}
# To get the sepset beliefs use the corresponding getters
In [15]: belief_propagation.get_sepset_beliefs()
Out[15]: {frozenset({('traffic_jam', 'long_queues'),
('traffic_jam', 'rain', 'accident')}): <Factor
representing phi(traffic_jam:2) at 0x7f565ee0def0>,
frozenset({('traffic_jam', 'late_for_school',
'getting_up_late'),
('traffic_jam', 'long_queues')}): <Factor representing phi(traffic_jam:2) at 0x7f565ee0dc18>}In the previous section, we saw how to compute the probability of variables present in the same cluster. Now, let's consider a situation where we want to compute the probability of both being late for school (L) and long queues (Q). These two variables are not present in the same cluster. So, to compute their probabilities, one naive approach would be to force our clique tree to have these two variables in the same cluster. However, this clique tree is not the optimal one, hence it would negate all the advantages of the belief propagation algorithm. The other approach is to perform variable elimination over the calibrated clique tree.
The algorithm for performing queries of variables not present in same cluster can be summarized as follows:
-
Select a subtree
 of the calibrated clique tree
of the calibrated clique tree  , such that the query variable
, such that the query variable  . Let
. Let  be a set of factors on which variable elimination is to be performed. Select a cluster of the clique tree
be a set of factors on which variable elimination is to be performed. Select a cluster of the clique tree  as the root node and add its belief to
as the root node and add its belief to  for each node in the clique tree
for each node in the clique tree  except the root node.
except the root node.

-
Add it to
 . Let Z be a random set of random variables present in
. Let Z be a random set of random variables present in  , except for the query variables. Perform variable elimination on the set of factors
, except for the query variables. Perform variable elimination on the set of factors  with respect to the variables Z.
with respect to the variables Z.
In pgmpy, this can be implemented as follows:
In [15]: belief_propagation.query(
variables=['no_of_people'],
evidence={'location': 1, 'quality': 1})
Out[15]: {'no_of_people': <Factor representing phi(no_of_people:2)
at 0x7f565ee0def0>Until now, we have been doing conditional probability queries only on the model. However, sometimes, rather than knowing the probability of some given states of variables, we might be interested in finding the states of the variables corresponding to the maximum probability in the joint distribution. This type of problem often arises when we want to predict the states of variables in our model, which is our general machine learning problem. So, let's take the example of our restaurant model. Let's assume that for some restaurant we know of, the quality is good, the cost is low, and the location is good, and we want to predict the number of people coming to the restaurant. In this case, rather than querying for the probabilities of states of the number of people, we would like to query for the state that has the highest probability, given that the quality is good, the cost is low, and the location is good. Similarly, in the case of speech recognition, given a signal, we are interested in finding the most likely utterance rather than the probability of individual phonemes.
Putting the MAP problem more formally, we are given a distribution ![]() defined by a set
defined by a set ![]() and an unnormalized
and an unnormalized ![]() , and we want to find an assignment
, and we want to find an assignment ![]() whose probability is at maximum:
whose probability is at maximum:

In the earlier equation, we used the unnormalized distribution to compute ![]() as it helps us avoid computing the full distribution, because computing the partition function Z requires all the values of the distribution. Overall, the MAP problem is to find the assignment
as it helps us avoid computing the full distribution, because computing the partition function Z requires all the values of the distribution. Overall, the MAP problem is to find the assignment ![]() for which
for which ![]() is at maximum.
is at maximum.
A number of algorithms have been proposed to find the most likely assignment. Most of these use local maximum assignments and graph structures to find the global maximum likely assignment.
We define the max-marginal of a function f relative to a set of variables Y as follows:
In simple words, ![]() returns the unnormalized probability value of the most likely assignment in
returns the unnormalized probability value of the most likely assignment in ![]() . Most of the algorithms work on first computing this set of local max-marginals, that is
. Most of the algorithms work on first computing this set of local max-marginals, that is ![]() , and then use this to compute the global maximum assignment, as we will see in the next sections.
, and then use this to compute the global maximum assignment, as we will see in the next sections.
