7
Filtering Rows
Introduction
Filtering data from a dataset is one of the most common and basic operations. There are numerous ways to filter (or subset) data in pandas with Boolean indexing. Boolean indexing (also known as Boolean selection) can be a confusing term, but in pandas-land, it refers to selecting rows by providing a Boolean array, a pandas Series with the same index, but a True or False for each row. The name comes from NumPy, where similar filtering logic works, so while it is really a Series with Boolean values in it, it is also referred to as a Boolean array.
We will begin by creating Boolean Series and calculating statistics on them and then move on to creating more complex conditionals before using Boolean indexing in a wide variety of ways to filter data.
Calculating Boolean statistics
It can be informative to calculate basic summary statistics on Boolean arrays. Each value of a Boolean array, the True or False, evaluates to 1 or 0 respectively, so all the Series methods that work with numerical values also work with Booleans.
In this recipe, we create a Boolean array by applying a condition to a column of data and then calculate summary statistics from it.
How to do it…
- Read in the movie dataset, set the index to the movie title, and inspect the first few rows of the
durationcolumn:>>> import pandas as pd >>> import numpy as np >>> movie = pd.read_csv( ... "data/movie.csv", index_col="movie_title" ... ) >>> movie[["duration"]].head() Duration movie_title Avatar 178.0 Pirates of the Caribbean: At World's End 169.0 Spectre 148.0 The Dark Knight Rises 164.0 Star Wars: Episode VII - The Force Awakens NaN - Determine whether the duration of each movie is longer than two hours by using the greater than comparison operator with the
durationcolumn:>>> movie_2_hours = movie["duration"] > 120 >>> movie_2_hours.head(10) movie_title Avatar True Pirates of the Caribbean: At World's End True Spectre True The Dark Knight Rises True Star Wars: Episode VII - The Force Awakens False John Carter True Spider-Man 3 True Tangled False Avengers: Age of Ultron True Harry Potter and the Half-Blood Prince True Name: duration, dtype: bool - We can now use this Series to determine the number of movies that are longer than two hours:
>>> movie_2_hours.sum() 1039 - To find the percentage of movies in the dataset longer than two hours, use the
.meanmethod:>>> movie_2_hours.mean() * 100 21.13506916192026 - Unfortunately, the output from step 4 is misleading. The duration column has a few missing values. If you look back at the DataFrame output from step 1, you will see that the last row is missing a value for
duration. The Boolean condition in step 2 returnsFalsefor this. We need to drop the missing values first, then evaluate the condition and take the mean:>>> movie["duration"].dropna().gt(120).mean() * 100 21.199755152009794 - Use the
.describemethod to output summary statistics on the Boolean array:>>> movie_2_hours.describe() count 4916 unique 2 top False freq 3877 Name: duration, dtype: object
How it works…
Most DataFrames will not have columns of Booleans like our movie dataset. The most straightforward method to produce a Boolean array is to apply a conditional operator to one of the columns. In step 2, we use the greater than comparison operator to test whether the duration of each movie was more than 120 minutes. Steps 3 and 4 calculate two important quantities from a Boolean Series, its sum and mean. These methods are possible as Python evaluates False and True as 0 and 1, respectively.
You can prove to yourself that the mean of a Boolean array represents the percentage of True values. To do this, use the .value_counts method to count with the normalize parameter set to True to get its distribution:
>>> movie_2_hours.value_counts(normalize=True)
False 0.788649
True 0.211351
Name: duration, dtype: float64
Step 5 alerts us to the incorrect result from step 4. Even though the duration column had missing values, the Boolean condition evaluated all these comparisons against missing values as False. Dropping these missing values allows us to calculate the correct statistic. This is done in one step through method chaining.
Important takeaway: You want to make sure you have dealt with missing values before making calculations!
Step 6 shows that pandas applies the .describe method to Boolean arrays the same way it applies it to a column of objects or strings, by displaying frequency information. This is a natural way to think about Boolean arrays, rather than displaying quantiles.
If you wanted quantile information, you could cast the Series into integers:
>>> movie_2_hours.astype(int).describe()
count 4916.000000
mean 0.211351
std 0.408308
min 0.000000
25% 0.000000
50% 0.000000
75% 0.000000
max 1.000000
Name: duration, dtype: float64
There's more…
It is possible to compare two columns from the same DataFrame to produce a Boolean Series. For instance, we could determine the percentage of movies that have actor 1 with more Facebook likes than actor 2. To do this, we would select both of these columns and then drop any of the rows that had missing values for either movie. Then we would make the comparison and calculate the mean:
>>> actors = movie[
... ["actor_1_facebook_likes", "actor_2_facebook_likes"]
... ].dropna()
>>> (
... actors["actor_1_facebook_likes"]
... > actors["actor_2_facebook_likes"]
... ).mean()
0.9777687130328371
Constructing multiple Boolean conditions
In Python, Boolean expressions use the built-in logical operators and, or, and not. These keywords do not work with Boolean indexing in pandas and are respectively replaced with &, |, and ~. Additionally, when combining expressions, each expression must be wrapped in parentheses, or an error will be raised (due to operator precedence).
Constructing a filter for your dataset might require combining multiple Boolean expressions together to pull out the rows you need. In this recipe, we construct multiple Boolean expressions before combining them to find all the movies that have an imdb_score greater than 8, a content_rating of PG-13, and a title_year either before 2000 or after 2009.
How to do it…
- Load in the movie dataset and set the title as the index:
>>> movie = pd.read_csv( ... "data/movie.csv", index_col="movie_title" ... ) - Create a variable to hold each filter as a Boolean array:
>>> criteria1 = movie.imdb_score > 8 >>> criteria2 = movie.content_rating == "PG-13" >>> criteria3 = (movie.title_year < 2000) | ( ... movie.title_year > 2009 ... ) - Combine all the filters into a single Boolean array:
>>> criteria_final = criteria1 & criteria2 & criteria3 >>> criteria_final.head() movie_title Avatar False Pirates of the Caribbean: At World's End False Spectre False The Dark Knight Rises True Star Wars: Episode VII - The Force Awakens False dtype: bool
How it works…
All values in a Series can be compared against a scalar value using the standard comparison operators (<, >, ==, !=, <=, and >=). The expression movie.imdb_score > 8 yields a Boolean array where all imdb_score values exceeding 8 are True and those less than or equal to 8 are False. The index of this Boolean array has the same index as the movie DataFrame.
The criteria3 variable is created by combining two Boolean arrays. Each expression must be enclosed in parentheses to function properly. The pipe character, |, is used to create a logical or condition between each of the values in both Series.
All three criteria need to be True to match the requirements of the recipe. They are each combined using the ampersand character, &, which creates a logical and condition between each Series value.
There's more…
A consequence of pandas using different syntax for the logical operators is that operator precedence is no longer the same. The comparison operators have a higher precedence than and, or, and not. However, the operators that pandas uses (the bitwise operators &, |, and ~) have a higher precedence than the comparison operators, hence the need for parentheses. An example can help clear this up. Take the following expression:
>>> 5 < 10 and 3 > 4
False
In the preceding expression, 5 < 10 evaluates first, followed by 3 > 4, and finally, the and evaluates. Python progresses through the expression as follows:
>>> 5 < 10 and 3 > 4
False
>>> True and 3 > 4
False
>>> True and False
False
>>> False
False
Let's take a look at what would happen if the expression in criteria3 was written as follows:
>>> movie.title_year < 2000 | movie.title_year > 2009
Traceback (most recent call last):
...
TypeError: ufunc 'bitwise_or' not supported for the input types, and the inputs could not be safely coerced to any supported types according to the casting rule ''safe''
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
...
TypeError: cannot compare a dtyped [float64] array with a scalar of type [bool]
As the bitwise operators have higher precedence than the comparison operators, 2000 | movie.title_year is evaluated first, which is nonsensical and raises an error. Therefore, we need parentheses to enforce operator precedence.
Why can't pandas use and, or, and not? When these keywords are evaluated, Python attempts to find the truthiness of the objects as a whole. As it does not make sense for a Series as a whole to be either True or False – only each element – pandas raises an error.
All objects in Python have a Boolean representation, which is often referred to as truthiness. For instance, all integers except 0 are considered True. All strings except the empty string are True. All non-empty sets, tuples, dictionaries, and lists are True. In general, to evaluate the truthiness of a Python object, pass it to the bool function. An empty DataFrame or Series does not evaluate as True or False, and instead, an error is raised.
Filtering with Boolean arrays
Both Series and DataFrame can be filtered with Boolean arrays. You can index this directly off of the object or off of the .loc attribute.
This recipe constructs two complex filters for different rows of movies. The first filters movies with an imdb_score greater than 8, a content_rating of PG-13, and a title_year either before 2000 or after 2009. The second filter consists of those with an imdb_score less than 5, a content_rating of R, and a title_year between 2000 and 2010. Finally, we will combine these filters.
How to do it…
- Read in the movie dataset, set the index to
movie_title, and create the first set of criteria:>>> movie = pd.read_csv( ... "data/movie.csv", index_col="movie_title" ... ) >>> crit_a1 = movie.imdb_score > 8 >>> crit_a2 = movie.content_rating == "PG-13" >>> crit_a3 = (movie.title_year < 2000) | ( ... movie.title_year > 2009 ... ) >>> final_crit_a = crit_a1 & crit_a2 & crit_a3 - Create criteria for the second set of movies:
>>> crit_b1 = movie.imdb_score < 5 >>> crit_b2 = movie.content_rating == "R" >>> crit_b3 = (movie.title_year >= 2000) & ( ... movie.title_year <= 2010 ... ) >>> final_crit_b = crit_b1 & crit_b2 & crit_b3 - Combine the two sets of criteria using the pandas or operator. This yields a Boolean array of all movies that are members of either set:
>>> final_crit_all = final_crit_a | final_crit_b >>> final_crit_all.head() movie_title Avatar False Pirates of the Caribbean: At World's End False Spectre False The Dark Knight Rises True Star Wars: Episode VII - The Force Awakens False dtype: bool - Once you have your Boolean array, you pass it to the index operator to filter the data:
>>> movie[final_crit_all].head() color ... movie/likes movie_title ... The Dark Knight Rises Color ... 164000 The Avengers Color ... 123000 Captain America: Civil War Color ... 72000 Guardians of the Galaxy Color ... 96000 Interstellar Color ... 349000 - We can also filter off of the
.locattribute:>>> movie.loc[final_crit_all].head() color ... movie/likes movie_title ... The Dark Knight Rises Color ... 164000 The Avengers Color ... 123000 Captain America: Civil War Color ... 72000 Guardians of the Galaxy Color ... 96000 Interstellar Color ... 349000 - In addition, we can specify columns to select with the
.locattribute:>>> cols = ["imdb_score", "content_rating", "title_year"] >>> movie_filtered = movie.loc[final_crit_all, cols] >>> movie_filtered.head(10) imdb_score content_rating title_year movie_title The Dark ... 8.5 PG-13 2012.0 The Avengers 8.1 PG-13 2012.0 Captain A... 8.2 PG-13 2016.0 Guardians... 8.1 PG-13 2014.0 Interstellar 8.6 PG-13 2014.0 Inception 8.8 PG-13 2010.0 The Martian 8.1 PG-13 2015.0 Town & Co... 4.4 R 2001.0 Sex and t... 4.3 R 2010.0 Rollerball 3.0 R 2002.0
How it works…
In step 1 and step 2, each set of criteria is built from simpler Boolean arrays. It is not necessary to create a different variable for each Boolean expression as done here, but it does make it far easier to read and debug any logic mistakes. As we desire both sets of movies, step 3 uses the pandas logical or operator to combine them.
In step 4, we pass the Series of Booleans created from step 3 directly to the index operator. Only the movies with True values from final_crit_all are selected.
Filtering also works with the .loc attribute, as seen in step 6, by simultaneously selecting rows (using the Boolean array) and columns. This slimmed DataFrame is far easier to check manually as to whether the logic was implemented correctly.
The .iloc attribute does not support Boolean arrays! If you pass in a Boolean Series to it, an exception will get raised. However, it does work with NumPy arrays, so if you call the .to_numpy() method, you can filter with it:
>>> movie.iloc[final_crit_all]
Traceback (most recent call last):
...
ValueError: iLocation based boolean indexing cannot use an indexable as a mask
>>> movie.iloc[final_crit_all.to_numpy()]
color ... movie/likes
movie_title ...
The Dark Knight Rises Color ... 164000
The Avengers Color ... 123000
Captain America: Civil War Color ... 72000
Guardians of the Galaxy Color ... 96000
Interstellar Color ... 349000
... ... ... ...
The Young Unknowns Color ... 4
Bled Color ... 128
Hoop Dreams Color ... 0
Death Calls Color ... 16
The Legend of God's Gun Color ... 13
There's more…
As was stated earlier, it is possible to use one long Boolean expression in place of several other shorter ones. To replicate the final_crit_a variable from step 1 with one long line of code, we can do the following:
>>> final_crit_a2 = (
... (movie.imdb_score > 8)
... & (movie.content_rating == "PG-13")
... & (
... (movie.title_year < 2000)
... | (movie.title_year > 2009)
... )
... )
>>> final_crit_a2.equals(final_crit_a)
True
Comparing row filtering and index filtering
It is possible to replicate specific cases of Boolean selection by taking advantage of the index.
In this recipe, we use the college dataset to select all institutions from a particular state with both Boolean indexing and index selection and then compare each of their performances against one another.
Personally, I prefer to filter by columns (using Boolean arrays) rather than on the index. Column filtering is more powerful as you can use other logical operators and filter on multiple columns.
How to do it…
- Read in the college dataset and use Boolean indexing to select all institutions from the state of Texas (
TX):>>> college = pd.read_csv("data/college.csv") >>> college[college["STABBR"] == "TX"].head() INSTNM ... GRAD_/_SUPP 3610 Abilene Christian University ... 25985 3611 Alvin Community College ... 6750 3612 Amarillo College ... 10950 3613 Angelina College ... PrivacySuppressed 3614 Angelo State University ... 21319.5 - To repeat this using index selection, move the
STABBRcolumn into the index. We can then use label-based selection with the.locindexer:>>> college2 = college.set_index("STABBR") >>> college2.loc["TX"].head() INSTNM ... GRAD_/_SUPP 3610 Abilene Christian University ... 25985 3611 Alvin Community College ... 6750 3612 Amarillo College ... 10950 3613 Angelina College ... PrivacySuppressed 3614 Angelo State University ... 21319.5 - Let's compare the speed of both methods:
>>> %timeit college[college['STABBR'] == 'TX'] 1.75 ms ± 187 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each) >>> %timeit college2.loc['TX'] 882 µs ± 69.3 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each) - Boolean indexing takes two times as long as index selection. As setting the index does not come for free, let's time that operation as well:
>>> %timeit college2 = college.set_index('STABBR') 2.01 ms ± 107 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
How it works…
Step 1 creates a Boolean Series by determining which rows of data have STABBR equal to TX. This Series is passed to the indexing operator, which selects the data. This process may be replicated by moving that same column to the index and using basic label-based index selection with .loc. Selection via the index is much faster than Boolean selection.
However, if you need to filter on multiple columns, you will have the overhead (and confusing code) from repeatedly switching the index. Again, my recommendation is not to switch the index, just to filter by it.
There's more…
This recipe only selects a single state. It is possible to select multiple states with both Boolean and index selection. Let's select Texas (TX), California (CA), and New York (NY). With Boolean selection, you can use the .isin method, but with indexing, just pass a list to .loc:
>>> states = ["TX", "CA", "NY"]
>>> college[college["STABBR"].isin(states)]
INSTNM CITY ... MD_EARN_WNE_P10 GRAD_DEBT_MDN_SUPP
192 Academy ... San Fran... ... 36000 35093
193 ITT Tech... Rancho C... ... 38800 25827.5
194 Academy ... Oakland ... NaN PrivacyS...
195 The Acad... Huntingt... ... 28400 9500
196 Avalon S... Alameda ... 21600 9860
... ... ... ... ... ...
7528 WestMed ... Merced ... NaN 15623.5
7529 Vantage ... El Paso ... NaN 9500
7530 SAE Inst... Emeryville ... NaN 9500
7533 Bay Area... San Jose ... NaN PrivacyS...
7534 Excel Le... San Antonio ... NaN 12125
>>> college2.loc[states]
INSTNM CITY ... MD_EARN_WNE_P10 GRAD_DEBT_MDN_SUPP
STABBR ...
TX Abilene ... Abilene ... 40200 25985
TX Alvin Co... Alvin ... 34500 6750
TX Amarillo... Amarillo ... 31700 10950
TX Angelina... Lufkin ... 26900 PrivacyS...
TX Angelo S... San Angelo ... 37700 21319.5
... ... ... ... ... ...
NY Briarcli... Patchogue ... 38200 28720.5
NY Jamestow... Salamanca ... NaN 12050
NY Pratt Ma... New York ... 40900 26691
NY Saint Jo... Patchogue ... 52000 22143.5
NY Franklin... Brooklyn ... 20000 PrivacyS...
There is quite a bit more to the story than what this recipe explains. pandas implements the index differently based on whether the index is unique or sorted. See the following recipe for more details.
Selecting with unique and sorted indexes
Index selection performance drastically improves when the index is unique or sorted. The prior recipe used an unsorted index that contained duplicates, which makes for relatively slow selections.
In this recipe, we use the college dataset to form unique or sorted indexes to increase the performance of index selection. We will continue to compare the performance to Boolean indexing as well.
If you are only selecting from a single column and that is a bottleneck for you, this recipe can save you ten times the effort
How to do it…
- Read in the college dataset, create a separate DataFrame with
STABBRas the index, and check whether the index is sorted:>>> college = pd.read_csv("data/college.csv") >>> college2 = college.set_index("STABBR") >>> college2.index.is_monotonic False - Sort the index from
college2and store it as another object:>>> college3 = college2.sort_index() >>> college3.index.is_monotonic True - Time the selection of the state of Texas (
TX) from all three DataFrames:>>> %timeit college[college['STABBR'] == 'TX'] 1.75 ms ± 187 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each) >>> %timeit college2.loc['TX'] 1.09 ms ± 232 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each) >>> %timeit college3.loc['TX'] 304 µs ± 17.8 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each) - The sorted index performs nearly an order of magnitude faster than Boolean selection. Let's now turn toward unique indexes. For this, we use the institution name as the index:
>>> college_unique = college.set_index("INSTNM") >>> college_unique.index.is_unique True - Let's select Stanford University with Boolean indexing. Note that this returns a DataFrame:
>>> college[college["INSTNM"] == "Stanford University"] INSTNM CITY ... MD_EARN_WNE_P10 GRAD_DEBT_MDN_SUPP 4217 Stanford... Stanford ... 86000 12782 - Let's select Stanford University with index selection. Note that this returns a Series:
>>> college_unique.loc["Stanford University"] CITY Stanford STABBR CA HBCU 0 MENONLY 0 WOMENONLY 0 ... PCTPELL 0.1556 PCTFLOAN 0.1256 UG25ABV 0.0401 MD_EARN_WNE_P10 86000 GRAD_DEBT_MDN_SUPP 12782 Name: Stanford University, Length: 26, dtype: object - If we want a DataFrame rather than a Series, we need to pass in a list of index values into
.loc:>>> college_unique.loc[["Stanford University"]] INSTNM CITY ... MD_EARN_WNE_P10 GRAD_DEBT_MDN_SUPP 4217 Stanford... Stanford ... 86000 12782 - They both produce the same data, just with different objects. Let's time each approach:
>>> %timeit college[college['INSTNM'] == 'Stanford University'] 1.92 ms ± 396 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each) >>> %timeit college_unique.loc[['Stanford University']] 988 µs ± 122 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
How it works…
When the index is not sorted and contains duplicates, as with college2, pandas will need to check every single value in the index to make the correct selection. When the index is sorted, as with college3, pandas takes advantage of an algorithm called binary search to improve search performance.
In the second half of the recipe, we use a unique column as the index. pandas implements unique indexes with a hash table, which makes for even faster selection. Each index location can be looked up in nearly the same time regardless of its length.
There's more…
Boolean selection gives much more flexibility than index selection as it is possible to condition on any number of columns. In this recipe, we used a single column as the index. It is possible to concatenate multiple columns together to form an index. For instance, in the following code, we set the index equal to the concatenation of the city and state columns:
>>> college.index = (
... college["CITY"] + ", " + college["STABBR"]
... )
>>> college = college.sort_index()
>>> college.head()
INSTNM CITY ... MD_EARN_WNE_P10 GRAD_DEBT_MDN_SUPP
ARTESIA, CA Angeles ... ARTESIA ... NaN 16850
Aberdeen, SD Presenta... Aberdeen ... 35900 25000
Aberdeen, SD Northern... Aberdeen ... 33600 24847
Aberdeen, WA Grays Ha... Aberdeen ... 27000 11490
Abilene, TX Hardin-S... Abilene ... 38700 25864
From here, we can select all colleges from a particular city and state combination without Boolean indexing. Let's select all colleges from Miami, FL:
>>> college.loc["Miami, FL"].head()
INSTNM CITY ... MD_EARN_WNE_P10 GRAD_DEBT_MDN_SUPP
Miami, FL New Prof... Miami ... 18700 8682
Miami, FL Manageme... Miami ... PrivacyS... 12182
Miami, FL Strayer ... Miami ... 49200 36173.5
Miami, FL Keiser U... Miami ... 29700 26063
Miami, FL George T... Miami ... 38600 PrivacyS...
We can compare the speed of this compound index selection with Boolean indexing. There is almost an order of magnitude difference:
>>> %%timeit
>>> crit1 = college["CITY"] == "Miami"
>>> crit2 = college["STABBR"] == "FL"
>>> college[crit1 & crit2]
3.05 ms ± 66.4 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
>>> %timeit college.loc['Miami, FL']
369 µs ± 130 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
Translating SQL WHERE clauses
Many pandas users will have experience of interacting with a database using Structured Query Language (SQL). SQL is a standard to define, manipulate, and control data stored in a database
SQL is an important language for data scientists to know. Much of the world's data is stored in databases that require SQL to retrieve and manipulate it SQL syntax is fairly simple and easy to learn. There are many different SQL implementations from companies such as Oracle, Microsoft, IBM, and more.
Within a SQL SELECT statement, the WHERE clause is very common and filters data. This recipe will write pandas code that is equivalent to a SQL query that selects a certain subset of the employee dataset.
Suppose we are given a task to find all the female employees who work in the police or fire departments who have a base salary of between 80 and 120 thousand dollars.
The following SQL statement would answer this query for us:
SELECT
UNIQUE_ID,
DEPARTMENT,
GENDER,
BASE_SALARY
FROM
EMPLOYEE
WHERE
DEPARTMENT IN ('Houston Police Department-HPD',
'Houston Fire Department (HFD)') AND
GENDER = 'Female' AND
BASE_SALARY BETWEEN 80000 AND 120000;
This recipe assumes that you have a dump of the EMPLOYEE database in a CSV file and that you want to replicate the above query using pandas.
How to do it…
- Read in the employee dataset as a DataFrame:
>>> employee = pd.read_csv("data/employee.csv") - Before filtering out the data, it is helpful to do some manual inspection of each of the filtered columns to know the exact values that will be used in the filter:
>>> employee.dtypes UNIQUE_ID int64 POSITION_TITLE object DEPARTMENT object BASE_SALARY float64 RACE object EMPLOYMENT_TYPE object GENDER object EMPLOYMENT_STATUS object HIRE_DATE object JOB_DATE object dtype: object >>> employee.DEPARTMENT.value_counts().head() Houston Police Department-HPD 638 Houston Fire Department (HFD) 384 Public Works & Engineering-PWE 343 Health & Human Services 110 Houston Airport System (HAS) 106 Name: DEPARTMENT, dtype: int64 >>> employee.GENDER.value_counts() Male 1397 Female 603 Name: GENDER, dtype: int64 >>> employee.BASE_SALARY.describe() count 1886.000000 mean 55767.931601 std 21693.706679 min 24960.000000 25% 40170.000000 50% 54461.000000 75% 66614.000000 max 275000.000000 Name: BASE_SALARY, dtype: float64 - Write a single statement for each of the criteria. Use the
isinmethod to test equality to one of many values:>>> depts = [ ... "Houston Police Department-HPD", ... "Houston Fire Department (HFD)", ... ] >>> criteria_dept = employee.DEPARTMENT.isin(depts) >>> criteria_gender = employee.GENDER == "Female" >>> criteria_sal = (employee.BASE_SALARY >= 80000) & ( ... employee.BASE_SALARY <= 120000 ... ) - Combine all the Boolean arrays:
>>> criteria_final = ( ... criteria_dept & criteria_gender & criteria_sal ... ) - Use Boolean indexing to select only the rows that meet the final criteria:
>>> select_columns = [ ... "UNIQUE_ID", ... "DEPARTMENT", ... "GENDER", ... "BASE_SALARY", ... ] >>> employee.loc[criteria_final, select_columns].head() UNIQUE_ID DEPARTMENT GENDER BASE_SALARY 61 61 Houston ... Female 96668.0 136 136 Houston ... Female 81239.0 367 367 Houston ... Female 86534.0 474 474 Houston ... Female 91181.0 513 513 Houston ... Female 81239.0
How it works…
Before any filtering is done, you will need to know the exact string names that you want to filter by. The .value_counts method is one way to get both the exact string name and number of occurrences of string values.
The .isin method is equivalent to the SQL IN operator and accepts a list of all possible values that you would like to keep. It is possible to use a series of OR conditions to replicate this expression, but it would not be as efficient or idiomatic.
The criteria for salary, criteria_sal, is formed by combining two simple inequality expressions. All the criteria are combined with the pandas and operator, &, to yield a single Boolean array as the filter.
There's more…
For many operations, pandas has multiple ways to do the same thing. In the preceding recipe, the criteria for salary uses two separate Boolean expressions. Similar to SQL, Series have a .between method, with the salary criteria equivalently written as follows. We will stick in an underscore in the hardcoded numbers to help with legibility:
''' {.sourceCode .pycon}
>>> criteria_sal = employee.BASE_SALARY.between(
... 80_000, 120_000
... )
'''
Another useful application of .isin is to provide a sequence of values automatically generated by some other pandas statements. This would avoid any manual investigating to find the exact string names to store in a list. Conversely, let's try to exclude the rows from the top five most frequently occurring departments:
>>> top_5_depts = employee.DEPARTMENT.value_counts().index[
... :5
... ]
>>> criteria = ~employee.DEPARTMENT.isin(top_5_depts)
>>> employee[criteria]
UNIQUE_ID POSITION_TITLE ... HIRE_DATE JOB_DATE
0 0 ASSISTAN... ... 2006-06-12 2012-10-13
1 1 LIBRARY ... ... 2000-07-19 2010-09-18
4 4 ELECTRICIAN ... 1989-06-19 1994-10-22
18 18 MAINTENA... ... 2008-12-29 2008-12-29
32 32 SENIOR A... ... 1991-02-11 2016-02-13
... ... ... ... ... ...
1976 1976 SENIOR S... ... 2015-07-20 2016-01-30
1983 1983 ADMINIST... ... 2006-10-16 2006-10-16
1985 1985 TRUCK DR... ... 2013-06-10 2015-08-01
1988 1988 SENIOR A... ... 2013-01-23 2013-03-02
1990 1990 BUILDING... ... 1995-10-14 2010-03-20
The SQL equivalent of this would be as follows:
SELECT *
FROM
EMPLOYEE
WHERE
DEPARTMENT not in
(
SELECT
DEPARTMENT
FROM ( SELECT
DEPARTMENT,
COUNT(1) as CT
FROM
EMPLOYEE
GROUP BY
DEPARTMENT
ORDER BY
CT DESC
LIMIT 5
) );
Notice the use of the pandas not operator, ~, which negates all Boolean values of a Series.
Improving the readability of Boolean indexing with the query method
Boolean indexing is not necessarily the most pleasant syntax to read or write, especially when using a single line to write a complex filter. pandas has an alternative string-based syntax through the DataFrame query method that can provide more clarity.
This recipe replicates the earlier recipe in this chapter, Translating SQL WHERE clauses, but instead takes advantage of the .query method of the DataFrame. The goal here is to filter the employee data for female employees from the police or fire departments who earn a salary of between 80 and 120 thousand dollars.
How to do it…
- Read in the employee data, assign the chosen departments, and import columns to variables:
>>> employee = pd.read_csv("data/employee.csv") >>> depts = [ ... "Houston Police Department-HPD", ... "Houston Fire Department (HFD)", ... ] >>> select_columns = [ ... "UNIQUE_ID", ... "DEPARTMENT", ... "GENDER", ... "BASE_SALARY", ... ] - Build the query string and execute the method. Note that the
.querymethod does not like triple quoted strings spanning multiple lines, hence the ugly concatenation:>>> qs = ( ... "DEPARTMENT in @depts " ... " and GENDER == 'Female' " ... " and 80000 <= BASE_SALARY <= 120000" ... ) >>> emp_filtered = employee.query(qs) >>> emp_filtered[select_columns].head() UNIQUE_ID DEPARTMENT GENDER BASE_SALARY 61 61 Houston ... Female 96668.0 136 136 Houston ... Female 81239.0 367 367 Houston ... Female 86534.0 474 474 Houston ... Female 91181.0 513 513 Houston ... Female 81239.0
How it works…
Strings passed to the .query method are going to look more like plain English than normal pandas code. It is possible to reference Python variables using the at symbol (@), as with depts. All DataFrame column names are available in the query namespace by referencing their names without extra quotes. If a string is needed, such as Female, inner quotes will need to wrap it.
Another nice feature of the query syntax is the ability to combine Boolean operators using and, or, and not.
There's more…
Instead of manually typing in a list of department names, we could have programmatically created it. For instance, if we wanted to find all the female employees who were not a member of the top 10 departments by frequency, we can run the following code:
>>> top10_depts = (
... employee.DEPARTMENT.value_counts()
... .index[:10]
... .tolist()
... )
>>> qs = "DEPARTMENT not in @top10_depts and GENDER == 'Female'"
>>> employee_filtered2 = employee.query(qs)
>>> employee_filtered2.head()
UNIQUE_ID POSITION_TITLE ... HIRE_DATE JOB_DATE
0 0 ASSISTAN... ... 2006-06-12 2012-10-13
73 73 ADMINIST... ... 2011-12-19 2013-11-23
96 96 ASSISTAN... ... 2013-06-10 2013-06-10
117 117 SENIOR A... ... 1998-03-20 2012-07-21
146 146 SENIOR S... ... 2014-03-17 2014-03-17
Preserving Series size with the .where method
When you filter with Boolean arrays, the resulting Series or DataFrame is typically smaller. The .where method preserves the size of your Series or DataFrame and either sets the values that don't meet the criteria to missing or replaces them with something else. Instead of dropping all these values, it is possible to keep them.
When you combine this functionality with the other parameter, you can create functionality similar to coalesce found in databases.
In this recipe, we pass the .where method Boolean conditions to put a floor and ceiling on the minimum and maximum number of Facebook likes for actor 1 in the movie dataset.
How to do it…
- Read the movie dataset, set the movie title as the index, and select all the values in the
actor_1_facebook_likescolumn that are not missing:>>> movie = pd.read_csv( ... "data/movie.csv", index_col="movie_title" ... ) >>> fb_likes = movie["actor_1_facebook_likes"].dropna() >>> fb_likes.head() movie_title Avatar 1000.0 Pirates of the Caribbean: At World's End 40000.0 Spectre 11000.0 The Dark Knight Rises 27000.0 Star Wars: Episode VII - The Force Awakens 131.0 Name: actor_1_facebook_likes, dtype: float64 - Let's use the
describemethod to get a sense of the distribution:>>> fb_likes.describe() count 4909.000000 mean 6494.488491 std 15106.986884 min 0.000000 25% 607.000000 50% 982.000000 75% 11000.000000 max 640000.000000 Name: actor_1_facebook_likes, dtype: float64 - Additionally, we may plot a histogram of this Series to visually inspect the distribution. The code below calls
plt.subplotsto specify the figure size, but is not needed in general:>>> import matplotlib.pyplot as plt >>> fig, ax = plt.subplots(figsize=(10, 8)) >>> fb_likes.hist(ax=ax) >>> fig.savefig( ... "c7-hist.png", dpi=300 ... )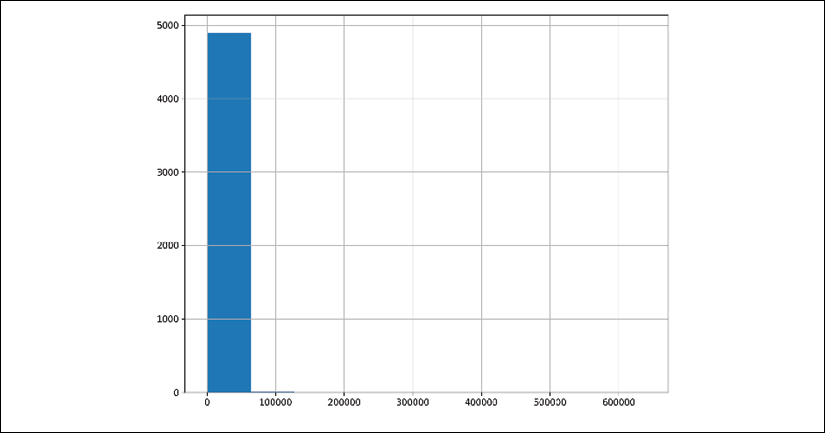
Default pandas histogram
- This visualization makes it difficult to get a sense of the distribution. On the other hand, the summary statistics from step 2 appear to be telling us that the data is highly skewed to the right with a few very large observations (more than an order of magnitude greater than the median). Let's create criteria to test whether the number of likes is fewer than 20,000:
>>> criteria_high = fb_likes < 20_000 >>> criteria_high.mean().round(2) 0.91 - About 91% of the movies have an actor 1 with fewer than 20,000 likes. We will now use the
.wheremethod, which accepts a Boolean array. The default behavior is to return a Series the same size as the original, but which has all theFalselocations replaced with a missing value:>>> fb_likes.where(criteria_high).head() movie_title Avatar 1000.0 Pirates of the Caribbean: At World's End NaN Spectre 11000.0 The Dark Knight Rises NaN Star Wars: Episode VII - The Force Awakens 131.0 Name: actor_1_facebook_likes, dtype: float64 - The second parameter to the
.wheremethod,other, allows you to control the replacement value. Let's change all the missing values to 20,000:>>> fb_likes.where(criteria_high, other=20000).head() movie_title Avatar 1000.0 Pirates of the Caribbean: At World's End 20000.0 Spectre 11000.0 The Dark Knight Rises 20000.0 Star Wars: Episode VII - The Force Awakens 131.0 Name: actor_1_facebook_likes, dtype: float64 - Similarly, we can create criteria to put a floor on the minimum number of likes. Here, we chain another
.wheremethod and replace the values not satisfying the condition to 300:>>> criteria_low = fb_likes > 300 >>> fb_likes_cap = fb_likes.where( ... criteria_high, other=20_000 ... ).where(criteria_low, 300) >>> fb_likes_cap.head() movie_title Avatar 1000.0 Pirates of the Caribbean: At World's End 20000.0 Spectre 11000.0 The Dark Knight Rises 20000.0 Star Wars: Episode VII - The Force Awakens 300.0 Name: actor_1_facebook_likes, dtype: float64 - The lengths of the original Series and the modified Series are the same:
>>> len(fb_likes), len(fb_likes_cap) (4909, 4909)
- Let's make a histogram with the modified Series. With the data in a much tighter range, it should produce a better plot:
>>> fig, ax = plt.subplots(figsize=(10, 8)) >>> fb_likes_cap.hist(ax=ax) >>> fig.savefig( ... "c7-hist2.png", dpi=300 ... )
A pandas histogram with a tighter range
How it works…
The .where method again preserves the size and shape of the calling object and does not modify the values where the passed Boolean is True. It was important to drop the missing values in step 1 as the .where method would have eventually replaced them with a valid number in future steps.
The summary statistics in step 2 give us some idea of where it would make sense to cap our data. The histogram from step 3, on the other hand, appears to clump all the data into one bin. The data has too many outliers for a plain histogram to make a good plot. The .where method allows us to place a ceiling and floor on our data, which results in a histogram with less variance.
There's more…
pandas actually has built-in methods, .clip, .clip_lower, and .clip_upper, that replicate this operation. The .clip method can set a floor and ceiling at the same time:
>>> fb_likes_cap2 = fb_likes.clip(lower=300, upper=20000)
>>> fb_likes_cap2.equals(fb_likes_cap)
True
Masking DataFrame rows
The .mask method performs the complement of the .where method. By default, it creates missing values wherever the Boolean condition is True. In essence, it is literally masking, or covering up, values in your dataset.
In this recipe, we will mask all rows of the movie dataset that were made after 2010 and then filter all the rows with missing values.
How to do it…
- Read the movie dataset, set the movie title as the index, and create the criteria:
>>> movie = pd.read_csv( ... "data/movie.csv", index_col="movie_title" ... ) >>> c1 = movie["title_year"] >= 2010 >>> c2 = movie["title_year"].isna() >>> criteria = c1 | c2 - Use the
.maskmethod on a DataFrame to remove the values for all the values in rows with movies that were made from 2010. Any movie that originally had a missing value fortitle_yearis also masked:>>> movie.mask(criteria).head() color ... movie_title ... Avatar Color ... Pirates of the Caribbean: At World's End Color ... Spectre NaN ... The Dark Knight Rises NaN ... Star Wars: Episode VII - The Force Awakens NaN ... - Notice how all the values in the third, fourth, and fifth rows from the preceding DataFrame are missing. Chain the
.dropnamethod to remove rows that have all values missing:>>> movie_mask = movie.mask(criteria).dropna(how="all") >>> movie_mask.head() color ... movie_title ... Avatar Color ... Pirates of the Caribbean: At World's End Color ... Spider-Man 3 Color ... Harry Potter and the Half-Blood Prince Color ... Superman Returns Color ... - The operation in step 3 is just a complex way of doing basic Boolean indexing. We can check whether the two methods produce the same DataFrame:
>>> movie_boolean = movie[movie["title_year"] < 2010] >>> movie_mask.equals(movie_boolean) False - The
.equalsmethod informs us that they are not equal. Something is wrong. Let's do some sanity checking and see whether they are the same shape:>>> movie_mask.shape == movie_boolean.shape True - When we used the preceding
.maskmethod, it created many missing values. Missing values arefloatdata types, so any column that was anintegertype that got missing values was converted to afloattype. The.equalsmethod returnsFalseif the data types of the columns are different, even if the values are the same. Let's check the equality of the data types to see whether this scenario happened:>>> movie_mask.dtypes == movie_boolean.dtypes color True director_name True num_critic_for_reviews True duration True director_facebook_likes True ... title_year True actor_2_facebook_likes True imdb_score True aspect_ratio True movie_facebook_likes False Length: 27, dtype: bool - It turns out that a couple of columns don't have the same data type. pandas has an alternative for these situations. In its
testingmodule, which is primarily used by developers, there is a function,assert_frame_equal, that allows you to check the equality of Series and DataFrames without also checking the equality of the data types:>>> from pandas.testing import assert_frame_equal >>> assert_frame_equal( ... movie_boolean, movie_mask, check_dtype=False ... )
How it works…
By default, the .mask method fills in rows where the Boolean array is True with NaN. The first parameter to the .mask method is a Boolean array. Because the .mask method is called from a DataFrame, all of the values in each row where the condition is True change to missing. Step 3 uses this masked DataFrame to drop the rows that contain all missing values. Step 4 shows how to do this same procedure with index operations.
During data analysis, it is important to continually validate results. Checking the equality of a Series and a DataFrame is one approach to validation. Our first attempt, in step 4, yielded an unexpected result. Some basic sanity checking, such as ensuring that the number of rows and columns are the same, or that the row and column names are the same, are good checks before going deeper.
Step 6 compares the data types of the two Series. It is here where we uncover the reason why the DataFrames were not equivalent. The .equals method checks that both the values and data types are the same. The assert_frame_equal function from step 7 has many available parameters to test equality in a variety of ways. Notice that there is no output after calling assert_frame_equal. This method returns None when two DataFrames are equal and raises an error when they are not.
There's more…
Let's compare the speed difference between masking and dropping missing rows and filtering with Boolean arrays. Filtering is about an order of magnitude faster in this case:
>>> %timeit movie.mask(criteria).dropna(how='all')
11.2 ms ± 144 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
>>> %timeit movie[movie['title_year'] < 2010]
1.07 ms ± 34.9 μs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
Selecting with Booleans, integer location, and labels
Previously, we covered a wide range of recipes on selecting different subsets of data through the .iloc and .loc attributes. Both of these select rows and columns simultaneously by either integer location or label.
In this recipe, we will filter both rows and columns with the .iloc and .loc attributes.
How to do it…
- Read in the movie dataset, set the index as the title, and then create a Boolean array matching all movies with a content rating of
Gand an IMDB score less than 4:>>> movie = pd.read_csv( ... "data/movie.csv", index_col="movie_title" ... ) >>> c1 = movie["content_rating"] == "G" >>> c2 = movie["imdb_score"] < 4 >>> criteria = c1 & c2 - Let's first pass these criteria to
.locto filter the rows:>>> movie_loc = movie.loc[criteria] >>> movie_loc.head() color ... movie/likes movie_title ... The True Story of Puss'N Boots Color ... 90 Doogal Color ... 346 Thomas and the Magic Railroad Color ... 663 Barney's Great Adventure Color ... 436 Justin Bieber: Never Say Never Color ... 62000 - Let's check whether this DataFrame is exactly equal to the one generated directly from the indexing operator:
>>> movie_loc.equals(movie[criteria]) True - Now, let's attempt the same Boolean indexing with the
.ilocindexer:>>> movie_iloc = movie.iloc[criteria] Traceback (most recent call last): ... ValueError: iLocation based boolean indexing cannot use an indexable as a mask - It turns out that we cannot directly use a Series of Booleans because of the index. We can, however, use an ndarray of Booleans. To get the array, use the
.to_numpy()method:>>> movie_iloc = movie.iloc[criteria.to_numpy()] >>> movie_iloc.equals(movie_loc) True - Although not very common, it is possible to do Boolean indexing to select particular columns. Here, we select all the columns that have a data type of 64-bit integers:
>>> criteria_col = movie.dtypes == np.int64 >>> criteria_col.head() color False director_name False num_critic_for_reviews False duration False director_facebook_likes False dtype: bool >>> movie.loc[:, criteria_col].head() num_voted_users cast_total_facebook_likes movie_facebook_likes movie_title Avatar 886204 4834 33000 Pirates o... 471220 48350 0 Spectre 275868 11700 85000 The Dark ... 1144337 106759 164000 Star Wars... 8 143 0 - As
criteria_colis a Series, which always has an index, you must use the underlying ndarray to make it work with.iloc. The following produces the same result as step 6:>>> movie.iloc[:, criteria_col.to_numpy()].head() num_voted_users cast_total_facebook_likes movie_facebook_likes movie_title Avatar 886204 4834 33000 Pirates o... 471220 48350 0 Spectre 275868 11700 85000 The Dark ... 1144337 106759 164000 Star Wars... 8 143 0 - When using
.loc,you can use a Boolean array to select rows, and specify the columns you want with a list of labels. Remember, you need to put a comma between the row and column selections. Let's keep the same row criteria and select thecontent_rating,imdb_score,title_year, andgrosscolumns:>>> cols = [ ... "content_rating", ... "imdb_score", ... "title_year", ... "gross", ... ] >>> movie.loc[criteria, cols].sort_values("imdb_score") content_rating imdb_score title_year gross movie_title Justin Bi... G 1.6 2011.0 73000942.0 Sunday Sc... G 2.5 2008.0 NaN Doogal G 2.8 2006.0 7382993.0 Barney's ... G 2.8 1998.0 11144518.0 The True ... G 2.9 2009.0 NaN Thomas an... G 3.6 2000.0 15911333.0 - You can create this same operation with
.iloc, but you need to specify the position of the columns:>>> col_index = [movie.columns.get_loc(col) for col in cols] >>> col_index [20, 24, 22, 8] >>> movie.iloc[criteria.to_numpy(), col_index].sort_values( ... "imdb_score" ... ) content_rating imdb_score title_year gross movie_title Justin Bi... G 1.6 2011.0 73000942.0 Sunday Sc... G 2.5 2008.0 NaN Doogal G 2.8 2006.0 7382993.0 Barney's ... G 2.8 1998.0 11144518.0 The True ... G 2.9 2009.0 NaN Thomas an... G 3.6 2000.0 15911333.0
How it works…
Both the .iloc and .loc attributes have some support filtering with Boolean arrays (with the caveat that .iloc cannot be passed a Series but the underlying ndarray.) Let's take a look at the one-dimensional ndarray underlying criteria:
>>> a = criteria.to_numpy()
>>> a[:5]
array([False, False, False, False, False])
>>> len(a), len(criteria)
(4916, 4916)
The array is the same length as the Series, which is the same length as the movie DataFrame. The integer location for the Boolean array aligns with the integer location of the DataFrame, and the filter happens as expected. These arrays also work with the .loc attribute as well, but they are a necessity with .iloc.
Steps 6 and 7 show how to filter by columns instead of by rows. The colon, :, is needed to indicate the selection of all the rows. The comma following the colon separates the row and column selections. However, there is actually a much easier way to select columns with integer data types and that is through the .select_dtypes method:
>>> movie.select_dtypes(int)
num_voted_users cast_total_facebook_likes
movie_title
Avatar 886204 4834
Pirates o... 471220 48350
Spectre 275868 11700
The Dark ... 1144337 106759
Star Wars... 8 143
... ... ...
Signed Se... 629 2283
The Follo... 73839 1753
A Plague ... 38 0
Shanghai ... 1255 2386
My Date w... 4285 163
Steps 8 and 9 show how to do row and column selections simultaneously. The rows were specified by a Boolean array and the columns were specified with a list of columns. You place a comma between the row and column selections. Step 9 uses a list comprehension to loop through all the desired column names to find their integer location with the index method .get_loc.
