- Extract the train dataset (trainX with all 784 independent variables and trainY with the respective 10 binary outputs):
trainX <- mnist$train$images
trainY <- mnist$train$labels
- Run a PCA on the trainX data:
PCA_model <- prcomp(trainX, retx=TRUE)
- Run an RBM on the trainX data:
RBM_model <- rbm(trainX, retx=TRUE, max_epoch=500,num_hidden =900)
- Predict on the train data using the generated models. In the case of the RBM model, generate probabilities:
PCA_pred_train <- predict(PCA_model)
RBM_pred_train <- predict(RBM_model,type='probs')
- Convert the outcomes into data frames:
PCA_pred_train <- as.data.frame(PCA_pred_train)
class="MsoSubtleEmphasis">RBM_pred_train <- as.data.frame(as.matrix(RBM_pred_train))
- Convert the 10-class binary trainY data frame into a numeric vector:
trainY_num<- as.numeric(stringi::stri_sub(colnames(as.data.frame(trainY))[max.col(as.data.frame(trainY),ties.method="first")],2))
- Plot the components generated using PCA. Here, the x-axis represents component 1 and the y-axis represents component 2. The following image shows the outcome of the PCA model:
ggplot(PCA_pred_train, aes(PC1, PC2))+
geom_point(aes(colour = trainY))+
theme_bw()+labs()+
theme(plot.title = element_text(hjust = 0.5))

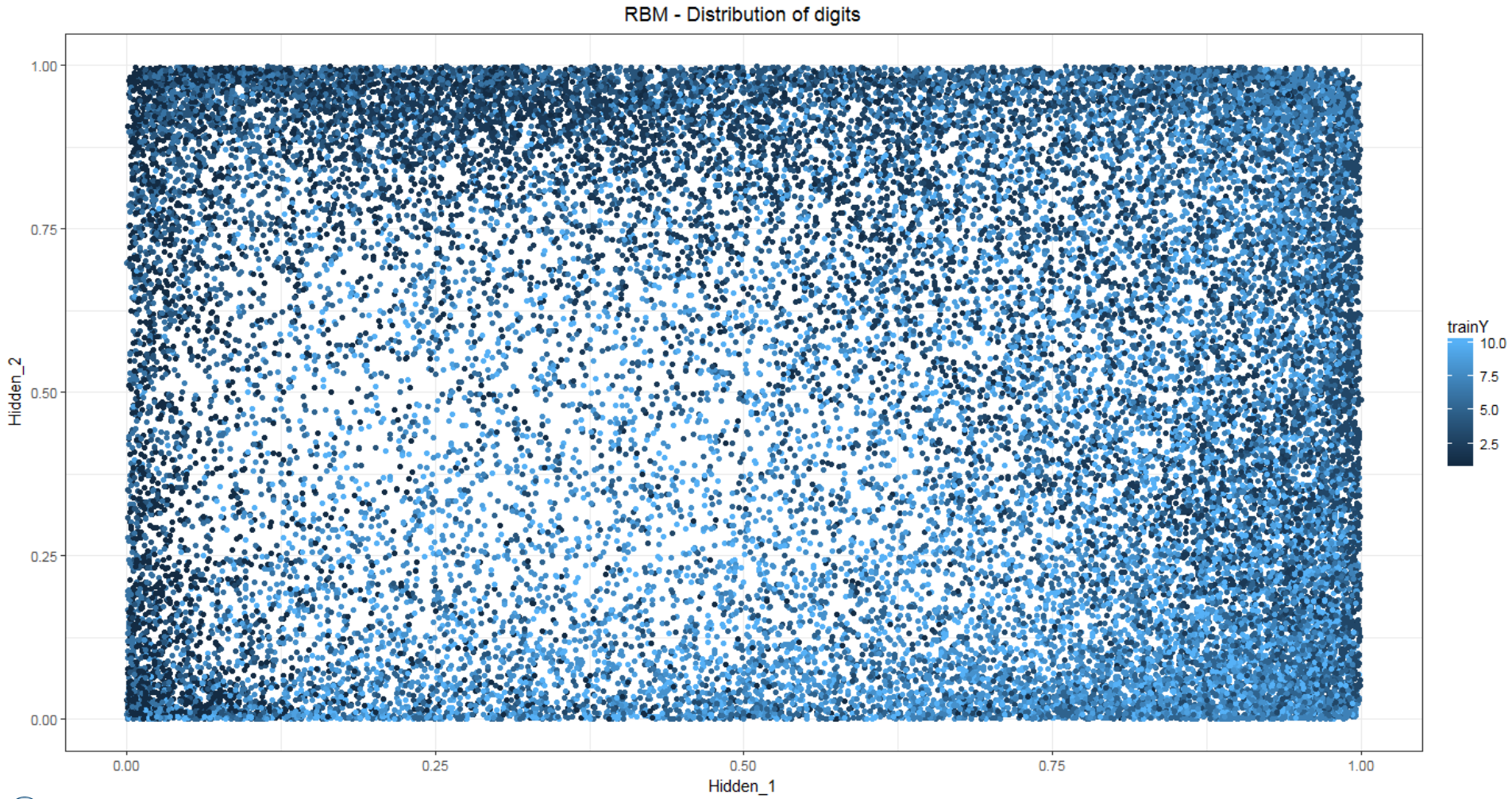
- Plot the hidden layers generated using PCA. Here, the x-axis represents hidden 1 and y-axis represents hidden 2. The following image shows the outcome of the RBM model:
ggplot(RBM_pred_train, aes(Hidden_2, Hidden_3))+
geom_point(aes(colour = trainY))+
theme_bw()+labs()+
theme(plot.title = element_text(hjust = 0.5))
The following code and image shows the cumulative variance explained by the principal components:
var_explain <- as.data.frame(PCA_model$sdev^2/sum(PCA_model$sdev^2))
var_explain <- cbind(c(1:784),var_explain,cumsum(var_explain[,1]))
colnames(var_explain) <- c("PcompNo.","Ind_Variance","Cum_Variance")
plot(var_explain$PcompNo.,var_explain$Cum_Variance, xlim = c(0,100),type='b',pch=16,xlab = "# of Principal Components",ylab = "Cumulative Variance",main = 'PCA - Explained variance')

The following code and image shows the decrease in the reconstruction training error while generating an RBM using multiple epochs:
plot(RBM_model,xlab = "# of epoch iterations",ylab = "Reconstruction error",main = 'RBM - Reconstruction Error')

