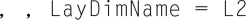The R Programming Language
You said, dear Descartes, that “je pense, donc je suis,”
Deriving existence from uncertainty.
Now, you are gone, and we say, “au revoir,”
Doubtless we think, René, therefore we R.1
In this book, you will learn how to actually do Bayesian data analysis. For any but the simplest models, that means using a computer. Because the computer results are so central to doing real Bayesian data analysis, examples of using the R computer programming language will be integrated into the simplest “toy” problems, so that R will not be an extra hurdle later.
The material in this chapter is rather dull reading because it basically amounts to a list (although a carefully scaffolded list) of basic commands in R along with illustrative examples. After reading the first few pages and nodding off, you may be tempted to skip ahead, and I wouldn't blame you. But much of the material in this chapter is crucial, and all of it will eventually be useful, so you should at least skim it all so you know where to return when the topics arise later.
Here are a few of the essential points of this chapter:
• Software installation. Section 3.1 describes how to install the R programming language. Section 3.1.1 describes how to install the R editor called RStudio. Section 3.2.1 describes how to install the programs written for this book.
• Data formats. Section 3.5.1 describes how to read data files into R. To understand the resulting format, you'll need to understand the data.frame structure from Section 3.4.4, which in turn requires understanding the list and matrix and factor structures (which is why those structures are explained before getting to the data.frame structure).
• Running programs. Section 3.7.2 explains how to run programs in R, and points out that it is important to set R's working directory to the folder in which the programs reside.
The R programming language is great at doing Bayesian statistics for a number of reasons. First, it's free! You can get it via the web and easily install it on your computer. Second, it's already a popular language for doing Bayesian statistics, so there are lots of resources available. Third, it is a powerful and easy, general-purpose computing language, so you can use it for many other applications too. The Bayesian MCMC packages that we will rely on later can be accessed from other programming environments but we'll use R. If you would like to learn more about the history of R, see its web page at http://www.r-project.org/ which also has extensive documentation about the R language.
3.1 Get the software
It's easy to get and install R, but there are a lot of optional details in the process, and the hardest part of installation is figuring out which little details do not apply to you!
Basic installation of R is easy. Go to http://cran.r-project.org/. (Web site addresses occasionally change. If the address stated here does not work, please search the web for “R language.” Be sure the site is legitimate before downloading anything to your computer.) At the top of that web page is a section headed “Download and Install R” followed by three links: Linux, MacOS, and Windows. These three links refer to the type of operating system used on your computer. Get the version that matches your computer's operating system. For example, if you click Windows, the next page that appears has a variety of links. The only one you need to click is “base,” which will get you to the most recent version of R for downloading. There are a few subsequent details specific to each operating system that you have to navigate on your own, but remember that centuries ago lots of people crossed various oceans in tiny wooden boats without any electronics, so you can navigate the small perils of R installation.2
3.1.1 A look at RStudio
The R programming language comes with its own basic user interface that is adequate for modest applications. But larger applications become unwieldy in the basic R user interface, and therefore it helps to install a more sophisticated R-friendly editor. There are a number of useful editors available, many of which are free, and they are constantly evolving. At the time of this writing, I recommend RStudio, which can be obtained from http://www.rstudio.com/. (Web site addresses occasionally change. If the address stated here does not work, please search the web for “RStudio.” Be sure the site is legitimate before downloading anything to your computer.) Just go to the web site, find the download link, and get the version appropriate to your computer operating system.
There are various ways to invoke R. One way is by starting R directly. For example, in Windows, you could just double-click the R icon, or find R in the list of programs from the Start menu. A second way to invoke R is via your favorite R editor. For example, invoke RStudio, which then automatically and transparently communicates with R. A third way to invoke R, and the best way for our purposes, is to associate files that have a “.R” extension on their filename with RStudio. Then, when the file is opened, your computer knows to invoke RStudio and open the file in RStudio. An example of doing this will be provided soon.
3.2 A simple example of R in action
Whether you are using the basic R interface or an editor such as RStudio, the primary window is a command-line interface. This window is constantly attentive to your every whim (well, every whim you can express in R). All you have to do is type in your wish and R will execute it as a command. The command line in R is marked by a prompt in the form of a greater-than sign: “>.” For example, if we want to know what 2 + 3 is, we can type it into R and R will provide the answer as text on a subsequent displayed line, like this:
Again, the “>“ symbol above indicates the R command prompt, at which we typed “2+3.” The next line above, “[1] 5,” shows R's reply. The answer, of course, is 5, but the line begins with a bracketed “[1]” to indicate that the first component of the answer is 5. As we will see later, variables in R are often multi-component structures, and therefore it can be informative to the user to display which component is being shown.
Throughout this book, programming commands are typeset in a distinctive font, like this, to distinguish them from English prose and to help demarcate the scope of the programming command when it is embedded in an English sentence.
A program, also known as a script, is just a list of commands that R executes. For example, you could first type in x=2 and then, as a second command, type in x+x, to which R will reply 4. This is because R assumes that when you type in x+x, you are really asking for the value of the sum of the value of x with the value of x, and you are not asking for an algebraic reformulation such as 2*x. We could save the list of commands as a program, or script, in a simple text file. Then we could run the program and R would step through the list of commands, in order. In R, running a script is called source-ing the script, because you are redirecting the source of commands from the command line to the script. To run a program stored in a file named MyProgram.R, we could type “source(“MyProgram.R”)” at R's command line, or click the “source” button in the R or RStudio menu. The example in the next section should help clarify.
As a simple example of what R can do, let's plot a quadratic function: y = x2. What looks like a smooth curve on a graph is actually a set of points connected by straight lines, but the lines are so small that the graph looks like a smooth curve. We must tell R where all those densely packed points should be positioned.
Every point is specified by its x and y coordinates, so we have to provide R with a list of x values and a list of corresponding y values. Let's arbitrarily select x values from −2 to +2, separated by intervals of 0.1. We have R set up the list of x values by using the built-in sequence function: x = seq( from = -2, to = 2, by = 0.1 ). Inside R, the variable x now refers to a list of 31 values: −2.0, −1.9, −1.8, …, +2.0. This sort of ordered list of numerical values is called a “vector” in R. We’ll discuss vectors and other structures in more detail later.
Next we tell R to create the corresponding y values. We type in y = xˆ2. R interprets “ˆ” to mean raising values to a power. Inside R, the variable y now refers to a vector of 41 values: 4.0, 3.61, 3.24, …, 4.0.
All that remains is telling R to make a plot of the x and y points, connected by lines. Conveniently, R has a built-in function called plot, which we call by entering plot (x , y , type=“l”). The segment of code, type=“l” (that's a letter “l“ not a numeral “1”) tells R to plot connecting lines between the points, with no distinctive symbols marking the points. If we omitted that part of the command, then R would plot only points by default, not the connecting lines. The resulting plot is shown in Figure 3.1, and the complete R code that generated the graph is shown below:

The command lines above include comments on each line to explain to the human reader what the line is intended to do. A comment in R begins with the “#”symbol, called a number sign or a pound sign. Anything on a line after the comment symbol is ignored by R.
3.2.1 Get the programs used with this book
The program above is stored as a file named SimpleGraph.R in the folder of programs that accompany the book. For the latest updates about programs used with this book, see the book's web site at https://sites.google.com/site/doingbayesiandataanalysis/, and the book's blog at http://doingbayesiandataanalysis.blogspot.com/. (Web site addresses occasionally change. If either of the addresses stated here does not work, please search the web for “Doing Bayesian Data Analysis” in quotes. Be sure the site is legitimate before downloading anything to your computer.) Specifically, find the link to the zip file that contains all the programs used with this book. Save the zip file at a convenient location on your computer where you normally save your data files and documents. (Do not save it in a protected area such as Programs in the Windows operating system.) Be sure to “unzip” the file or “extract all” of the programs from the zipped folder.
The simplest way to open the program is to associate the file with RStudio and then open the file. For example, in the Windows operating system, find the file SimpleGraph.R. Right-click it and select “Open with…” and then browse to RStudio. Be sure to select (i.e., check) the box that says to always use RStudio to open files of this type. Then, the file will open in the RStudio editor, ready for running and additional editing as desired.
One of the benefits of opening a program directly and letting it automatically invoke RStudio, instead of opening RStudio and subsequently loading the program, is that RStudio sets the working directory to the folder in which the program resides. The working directory is where R first looks for auxiliary programs and where R saves output files. If you invoke RStudio from scratch, instead of via a program file, then the working directory is a default generic folder and consequently R will not know where to find auxiliary programs used with this book. More information about setting the working directory will appear later.
3.3 Basic commands and operators in R
The rest of this chapter describes some of the important features of the R programming language. There are many other sources available online, including the introductory manual, the latest version of which should be available at this address: http://cran.r-project.org/doc/manuals/r-release/R-intro.pdf. If that link no longer works by the time you read this, do a little searching of documentation on the R home page or on the web.
3.3.1 Getting help in R
The plot function has many optional details that you can specify, such as the axis limits and labels, font sizes, etc. You can learn more about those details by getting help from R. Type the command ?”plot” and you can read all about it. In particular, the information directs you to another command, par, that controls all the plot parameters. To learn about it, type ?”par”. In general, it actually is helpful to use R's built-in help. To get a list of all sorts of online documentation, much of it written in readable prose instead of telegraphic lists, type help.start() including the empty parentheses. In particular, the list of manuals displayed by help.start() includes An Introduction to R, which is highly recommended. Another useful way to find help with R is through web search. In your favorite web searcher, type in the R terms you want help with.
Also very useful is the double question mark, which is an abbreviation for the help.search function, and which searches the entire help database for entries that contain the specified word or phrase. While ?”plot” returns the single help page that explains the plot function, ??”plot” returns a list of dozens of help pages that contain the word “plot.” (For more details, you can type ?”?” and ?”??” at R's command prompt.)
A highly recommended resource is a summary of basic R commands that can be found on a compact list available at this address: http://cran.r-project.org/doc/contrib/Short-refcard.pdf Other versions of reference cards can be found by searching the web with the phrase “R reference card.”
Much of the time you'll learn about features of R on an as-needed basis, and usually that means you'll look for examples of the sort of thing you want to do and then imitate the example. (Or, the example might at least provoke you into realizing that there is a better way to do it than the method in the example!) Therefore, most of the examples in this book have their full R code included. Hopefully, it will help you to study those examples as needed.
If you are already familiar with the programming languages Matlab or Python, you can find thesauruses of synonymous commands in R at this Web site: http://mathesaurus.sourceforge.net/.
3.3.2 Arithmetic and logical operators
The four arithmetic operators are typed as +, -, *, and /. Power is typed as ˆ. When operators are specified successively, they are processed left to right, except that power has precedence over multiplication and division, which have precedence over addition and subtraction. Make sure you understand the output returned by each of the following examples:
In general, it's a good idea to include explicit parentheses to be sure that the operations are carried out in the order that you intended. Details about operator precedence can be found in R by typing ?”Syntax”.
R also deals with the logical values TRUE and FALSE, along with logical operations of negation, !, conjunction, &, and disjunction, |. Negation has precedence, followed by conjunction then disjunction. For example,
3.3.3 Assignment, relational operators, and tests of equality
We can assign values to named variables. For example, x = 1 commands R to assign the value 1 to a variable named x. There is a synonymous syntax for the assignment operator that looks like an arrow: x <- 1. Originally, R only used the arrow-like symbol to indicate assignment, but in recent years, due to popular demand from users of other programming languages, R also allows the equal sign. This book will usually use the equal sign for assignment.
Purists avoid using the single equal sign for assignment, because it can be confused with the meaning(s) of the mathematical equal sign. In particular, it is perfectly valid in R to state x = x + 1, even though it does not make much sense in ordinary mathematical notation to say x = x + 1 (because that implies 0 = 1, which is false in ordinary arithmetic). In R, “x = x + 1” means to add 1 to the current value of x and assign the result to the variable named x. The new value of x replaces the old value of x. For example, consider the following sequence of R commands:
In practice, this possible misinterpretation of an R command as a mathematical statement of equality is rare or nonexistent. Besides, R uses the single equal sign for assignmentlike operations in arguments of functions and in definitions of list structures (as will be explained subsequently). Therefore, I am not deterred from using the single equal sign for assignment.
Beware, however, not be confuse the assignment operator = with a test for equality, which is denoted in R by a double equal sign. The double equal sign tests for equality and returns a value of TRUE or FALSE. For example:
There are also relational operators for inequalities:
If you use <- for the assignment operator instead of =, be certain not to accidentally insert a space: The expression x <- 3 assigns the value 3 to x, but the expression x < - 3 tests whether x is less than the value −3 and returns TRUE or FALSE (or an error message if x does not exist).
The double equal sign for testing equality, used above, is brittle and can give unintended results due to the limited precision of representing numbers in the computer's memory. For example, in most computers, the value of 0.5 − 0.3 does not equal the value of 0.3 − 0.1, even though mathematically they are equivalent. Therefore, R has another function, all.equal, for testing equality up to the degree of precision for the computer being used. For example:
Therefore, it's safe practice to use all.equal instead of == when testing equality, especially for nonlogical or noninteger values.
3.4 Variable types
All objects in R have a class which can indicate the structure and type of content in the object, and which can be recognized by some functions so that the functions treat different classes of objects different ways. For example, objects can be of class matrix, array, list, factor, data.frame, etc. The summary function, described later, detects the class of its argument and will summarize a numeric vector differently than a factor. In this section, a few important classes of objects are described.
3.4.1 Vector
A vector is simply an ordered list of elements of the same type. A vector is not technically a class in R, but a vector is the fundamental data structure. For example, a vector could be an ordered list of numbers like this, 〈2.718, 3.14,1.414〉, or an ordered list of strings like this, 〈“now”, “is”, “the”, “time”〉, or an ordered list of logical values like this, 〈TRUE, FALSE, FALSE, TRUE〉. By “ordered” I mean that R knows the elements by their position in the list, not that the elements themselves somehow go in increasing or decreasing value. For example, R knows that the third element of the vector 〈2.718, 3.14, 1.414〉 is 1.414 because the elements are ordered.
3.4.1.1 The combine function
The combine function, c (yes, the single letter “c”),makes vectors. Actually,the combine function can combine different types of data structures, but at this point we consider only its ability to create vectors by combining elements. For example, c( 2.718 , 3.14 , 1.414 ) combines the three numbers into a vector. We could then assign that vector to a variable named “x” by the command x = c( 2.718 , 3.14 , 1.414 ).
3.4.1.2 Component-by-component vector operations
R defaults to component-by-component vector operations. In particular, if x and y are two vectors of the same length, then x*y is the vector consisting of the products of corresponding components. For example:
R automatically applies a scalar operation to all elements of a vector. For example:
3.4.1.3 The colon operator and sequence function
The colon operator, :, makes sequences of integers. For example, 4:7 creates the vector 〈4, 5, 6, 7〉. The combine function and the colon operator are used very often in R programming. The colon operator has precedence over basic arithmetical operators, but not over the power operator. Consider carefully the output of these examples:
In general, to be sure that the computations are executed in the order that you intend, include explicit parentheses.
The sequence function, seq, is very handy for creating vectors that consist of regular sequences of numbers. In its basic form, the user specifies the starting value of the sequence, the ending value, and the increment between successive values, like this:
If the increment is not an exact divisor of the distance between the starting and ending values, the sequence will not exceed the ending value. For example:
The seq function is clever and will infer whatever value is omitted by the user. This capability can be very useful, for example if we want a sequence of a certain length and do not care about the exact end point or the increment. Consider these examples:
3.4.1.4 The replicate function
The replicate function, rep, is also very useful for creating vectors. Here are some examples:
Notice above that rep has three optional arguments after the first argument (which is the structure to be replicated). When only one integer is entered, as in the second line above, then rep assumes it is the number of times that the entire structure should be replicated. According to the R help file, “Normally just one of the additional arguments is specified, but if each is specified with either of the other two, its replication is performed first, and then that implied by times or length.out.” If you intend that each component should be replicated first, be sure to use the each argument explicitly, or use the times argument in its vector form.
Here is an example to test your understanding of the rep function:
What happened in that example is that the each=2 argument was applied first, thereby creating (internally) the vector c(“A”,”A”,”B”,”B”,”C”,”C”), which has six components. After that, the times=c(1,2,3,1,2,3) argument was applied, which created one copy of the first “A,” followed by two copies of the second “A,” followed by three copies of the first “B,” and so forth. In other words, the same result could be created by the following nested application of rep:
3.4.1.5 Getting at elements of a vector
Yoohoo, time to wake up! The methods of this section will be used often. In R, we can get at elements of a vector by referencing their position in the vector. There are three ways to reference elements: by numerical position, by logical inclusion, and by name. All three ways refer to positions by using square brackets after the vector name, like this: x[ ... ]. What differs between the methods is what goes between the square brackets.
We can refer to elements by their numerical position. For example, suppose we define a vector x=c( 2.718 , 3.14 , 1.414 , 47405 ). We can get the elements of the vector by referring to the rank positions inside square brackets after the vector name. Thus, x[c(2,4)] returns the second and fourth elements, as the vector 〈3.14, 47405〉. Another way to get at elements is by telling R which position not to include, by using a negative sign. For example, if you want all the elements except the first and third, you can type x[c(-1, -3)].
Another way to access elements of a vector is by a sequence of logical true and false values that specify whether or not to return the corresponding element. For example, x[c(FALSE, TRUE, FALSE, TRUE)] also returns the vector 〈3.14, 47405〉.
The elements of a vector can also, optionally, have names. For example, we can name the elements of the vector x with the command, names (x)=c( “e” , “pi” , “sqrt2” , “zipcode” ). Notice that the names are in quotes. Then we get at the components of the vector by specifying their names in square brackets. For example, x[c(“pi”, ”zipcode”)] returns the vector 〈3.14,47405〉 along with the names of those components.
To recapitulate and summarize, here are some different ways to get at the elements of a vector:
3.4.2 Factor
Factors are a type of vector in R for which the elements are categorical values that could also be ordered. The values are stored internally as integers with labeled levels. The best way to explain factors is by example.
Suppose we have data regarding the socio-economic status of five people, coded as one of “low,” “medium,” or “high.” Here are the data, in the vector named x:
The terms “high,” “medium,” and “low” get a bit unwieldy in large data sets, and therefore it can be useful to recode them into categorical indices, such as “1” for “low,” “2” for “medium,” and “3” for “high.” The data become a vector of indices along with a legend from translating from the indices to names for the indices. The resulting structure is called a “factor.” In R, the factor function converts the vector x to a factor:
To see what the factor xf looks like, we can type “xf” at R's command line and see what R returns:
Notice that R has extracted “levels” from the vector and listed them after the contents of the factor. The factor function read the contents of vector x and kept track of all the distinct elements, calling them the levels of the factor. By default, it ordered the levels alphabetically. It then recoded the contents of the vector in terms of the integer indices of the alphabetized levels. Thus, the original element “high” becomes integer “1” because “high” is alphabetically first among the three levels. You can think of the real contents of the factor as being the integer indices of the levels, along with a legend that decodes each integer into a level name. To see the integer indices of the levels explicitly, we can ask R to show us the factor as a numeric vector:
The factor command extracts the levels and orders them alphabetically by default. Unfortunately, alphabetical order depends on local language customs (e.g., uppercase letters might go before lowercase letters or after), and alphabetical order might not be the most meaningful order for a given application. If we want to specify a particular order for the levels, we can do so with additional arguments in the factor function, like this:
Notice now that the levels are displayed with less-than signs to indicate that they are ordered. And, when we examine the integer indices internal to the factor, they are coded according to the desired order, such that the first element, “high,” is coded as integer 3, not as 1:
It is good practice to explicitly specify the levels and their ordering when making a factor.
Sometimes we will want to reorder the levels of a factor that has already been created. This can happen, for example, when reading in a data file using default settings, but subsequently reordering to make the levels more meaningful. To reorder, just use the factor function again, explicitly indicating the desired levels and order. For example, we create factor xf as before:
But now we reorder the levels by applying the factor function to the existing factor xf:
Notice that explicitly reordering the levels has also changed the underlying integer coding of the elements in the vector, as it should. It is good practice to specify the ordering of the levels explicitly.
We might want to relabel the levels of a factor. For example, the vector x used elements named low, medium, and high. The levels argument of the factor function must refer to those exact terms for reading in the data. But for the resulting output, the levels can be relabeled to whatever might be more meaningful for the application, such as “Bottom SES,” “Middle SES,” and “Top SES.” We accomplish the relabeling in R with the labels argument:
Notice that the original data terms (low, medium, high) are no longer accessible in the factor. It is important to realize that relabeling the levels does not change their order or their integer coding. For example, if we specify labels=c(“Right”,”Left”,“UpsideDown”), the integers will be unchanged, but their labeling will no longer be meaningful.
Factors will appear frequently when R reads data from files. Factors can be very useful for some functions in R. But factors can be confusing if you are not aware that a variable is being stored by R as a factor and not as a vector. Factors can also be confusing if the default ordering of their levels is not intuitive.
3.4.3 Matrix and array
A matrix is simply a two-dimensional array of values of the same type. A matrix can be created in R using the matrix command. The first argument specifies the contents of the matrix, in order. Other arguments specify the size of the matrix, the order in which the matrix should be filled, and, optionally, the names of the dimensions and rows and columns. (The names are specified as a list, which is a structure that is described explicitly very soon, in Section 3.4.4.) Please note the comments in the following examples:
In the final example above, the “+” symbols at the beginning of some command lines are merely R's way of displaying that those lines are continuations of the same command, instead of new commands. You do not type in the plus symbols when you enter the command, just as you do not type in the greater-than symbol at the beginning of a command.
Just as with vectors, the components of matrices can be accessed via their indices or their names. The following shows two ways to access the element in the second row and third column of the matrix x:
As you may have inferred by now, the indices are ordered such that the first index refers to the row and the second index refers to the column.
An entire row or column of a matrix can be accessed by specifying its entire range or by leaving its range unspecified. For example:
Be very careful to include the comma when specifying a row or column of a matrix. If you use a numerical index for a row or column and accidentally leave out the comma, R will not complain, but will instead return the element at that position in the ordered contents. Consider these examples:
Notice in the final example above, when there was no comma, R returned the second element in the contents of the matrix. Although it is perfectly valid to refer to the elements of a matrix this way, without using row and column referents, it should usually be avoided unless there is some specific application that is made more efficient.
An array is a generalization of a matrix to multiple dimensions. There is really no need for a separate matrix function because it is merely the two-dimensional case of the array function. In the array function, the first argument specifies the ordered contents, the second argument specifies the size of each dimension, and an optional third argument specifies the names of the dimensions and levels within dimensions. (The names are specified as a list, which is a structure that is described explicitly very soon, in Section 3.4.4.) It is important to understand that the array function fills the array by incrementing the first index (row) first, then incrementing the second index (column) next, then incrementing the third index (layer) next, and so forth. Unlike the matrix function, there is no built-in way to load the contents into the array in a different ordering of dimensions.
Here is an example of a three-dimensional array. I have referred to the third dimension as a “layer.” Notice that the contents are the integers 1-24, and they are filled into the array by first incrementing the row, then the column, and then the layer.
3.4.4 List and data frame
The list structure is a generic vector in which components can be of different types, and named. The list structure was used in previous examples to specify dimension names in the matrix and array functions. Below is an example of a list in which the first element is a vector of integers named “a,” the second element is a matrix of integers named “b,” and the third element is a string named “c.”
The named components of a list can be referred to by appending the list name with a “$” and the component name. Consider these examples:
The contents of the list can also be accessed with (numerical) indices inside square brackets. There is an additional layer of nuance for referring to named components, however. We can get element i from a list, including its name, by putting i inside single square brackets. We can get the contents of element i by putting i inside double square brackets. Consider these examples:
A data frame is much like a matrix, insofar as it has several columns of equal length. But each column can be of a different type, and, in particular, columns can be factors. A data frame is redly a type of list in which each component is thought of as a named column of a matrix, with different columns possibly of different types. The elements of a data frame can be accessed as if it were a list or as if it were a matrix. Consider this example:
In the display of data frame d, above, the column of numbers on the far left side shows the row names supplied, by default, by the data.frame function. Do not confuse those row names with the contents of the columns.
The elements of the data frame can be accessed as for a list, by using names or single brackets or double brackets:
The elements of the data frame can also be accessed as for a matrix, using row and column indices:
Data frames are important because they are the default format for data loaded into R by the often-used function read.table, as we will explore next.
3.5 Loading and saving data
This book uses a typical convention for arranging data in which every row of a data file contains one measurement instance, which might be one person in a survey, or one trial in a response-time experiment. Each row contains the value of the key measurement to be explained or predicted, and each row also contains values that indicate predictors or explanatory variables for that instance. Examples, shown below, will clarify.
3.5.1 The read.csv and read.table functions
Consider a small sample of people, from whom we record their gender, hair color, and ask a random number between 1 and 10. We also record each person's first name, and which of two groups they will be assigned to. It is typical to save this sort of data as a computer file in a format called comma separated values,or CSV format. In CSV format, there is a column for each type of measurement, and a row for each person (or item) measured. The first row specifies the column names, and the information in each column is separated by commas. Here is an example:
CSV files are especially useful because they are generic text files that virtually any computer system can read. A disadvantage of CSV files is that they can take up a lot of memory space, but these days computer memory is cheap and plentiful.
CSV files are easily loaded into R's memory using the read.csv function. Suppose that the data above are saved in a file called HGN.csv. Then the data can be loaded into a variable I’ve named HGNdf as follows:
The resulting variable, HGNdf, is a data frame in R. Thus, the columns of HGNdf are vectors or factors, named according to the words in the first row of the CSV file, and all of length equal to the number of data rows in the CSV file.
It is important to note that columns with any character (non-numeric) entries are turned into factors (recall that Section 3.4.2 described factors). For example, the Hair column is a factor:
The levels of the factor are alphabetical by default. We might want to reorder the levels to be more meaningful. For example, in this case we might want to reorder the hair colors from lightest to darkest. We can do that after loading the data, like this:
There might be times when we do not want a column with character entries to be treated as a factor. For example, the Name column is treated by read.csv as a factor because it has character entries:
Because the names are never repeated, there are as many factor levels as entries in the column, and there might be little use in structuring it as a factor. To convert a factor to an ordinary vector, use the function as.vector, like this:
There might be times when a column of integers is read by read.csv as a numeric vector, when you intended it to be treated as indexical levels for grouping the data. In other words, you would like the column to be treated as a factor, not as a numeric vector. This happened in the present example with the Group column:
Notice above that no levels are associated with the Group column. It is easy to convert the column to a factor:
The read.csv function is a special case of the more general read.table function. You can learn more about it by typing ?”read.table” at R's command prompt. For example, you can turn off the default action of making character columns into factors.
3.5.2 Saving data from R
In most real research situations, data are obtained from some measurement device or survey outside of R. The collected data are then loaded into R using a function such as read.csv, as explained above. Sometimes, however, we want to save reformatted or transformed data from R.
One way to save a data table is with the write.csv function. For example, the command
saves the data frame HGNdf as the file named HGN.csv, without including the row names and without putting all character elements inside double quotes (which write.csv would otherwise do by default). It is important to understand that the resulting file loses all information about levels in factors, because it is only a raw text file with no summary information.
If you want to save data frames with all the factor information intact, then you can use the save command. The resulting file is in a special R format, not generic text, and the standard filename extension for this type of file is “.Rdata.” For example, the command
saves the data frame HGNdf as the file named HGN.Rdata with all its factor information, and with the name of data frame (i.e., “HGNdf”). If desired, several different variables can all be saved in a single Rdata file, simply by specifying them as the initial comma-separated arguments in the save command. All the variables are saved along with their names.
To retrieve an Rdata file, use the load function:
The load function executes without explicitly showing the user any changes in R's internal state. But R has, in fact, loaded the variables into its working memory. You can see the objects that R has in its active memory by typing
The output from the objects function shows that R has HGNdf in its memory, in this case because it loaded that variable from the file HGN.Rdata. In the editor RStudio, a convenient way to see what objects are in R's memory is by looking at the Workspace window (usually in the upper right or RStudio's display). The Workspace window shows all the objects, organized by type.
3.6 Some utility functions
A function is a process that takes some input, called the arguments, and produces some output, called the value. This section reviews some useful utility functions in R.
The summary function detects the type, or “class,” of the argument provided to it, and returns a summary appropriate for that class of object. For example, if we construct a vector consisting of numerical values, summary provides the minimum value in the vector, median value, etc., as shown by this example:
However, if we convert the vector to a factor, then the summary function provides a table with the frequency of each level:
If you put a data frame into the summary function, it will provide a summary of each column appropriate to the class of information in each column.
Other useful functions for examining data objects are head, tail, and str. The head function returns the first few components of the variable put in its argument. The str function returns a compact display of the structure of its argument. Type ?”head” (etc.) in R to learn more.
The aggregate function is very useful for summarizing data according to factor characteristics. To illustrate, recall the hair-gender-number data from Section 3.5.1, in which each person provided a self-generated number between 1 and 10. The data were read into a data frame named HGNdf, with the Group column converted to a factor. Suppose we want to know the median value of the numbers selected by each gender within each hair color. The aggregate function provides the answer. The first argument, specified as “x=,” is the data we want summarized. The “by=” argument is a list of factors for grouping the data. The “FUN=” argument is the function that should be applied to the groups of data. For example:
Notice that the names of the columns in the output were given defaults that are unrelated to the original variable names. If we want more meaningful output, we have to explicitly name the variables:
Alternatively, we can use the formula format for the arguments. In formula format, the first argument is a formula that uses the column names from the data frame to express what we want aggregated. For example, to specify that we want the Number aggregated by Group and Gender, we would use the formula Number ∼ Group + Gender. Butwe also have to tell the function what data frame we are referring to, and this is done with the data argument. Thus,
The formula format is used by many functions in R that take data frames as arguments. The aggregate function has several other arguments, and it can also be used to summarize time series. Of course, you can type ?“aggregate” for more information.
The formula format is used by many functions in R that take data frames as arguments. The aggregate function has several other arguments, and it can also be used to summarize time series. Of course, you can type ?“aggregate” for more information.
The aggregate function is also useful for counting how many times various levels of a factor occur, or how many times combinations of factor levels occur. For example, suppose we would like to know how many times each hair color occurs for each gender. We use the aggregate function by applying the sum function, aggregating on the gender and hair-color factors. But what value do we sum? We want to sum the number of rows of each combination, so we create a new column that contains all 1s, to explicitly indicate that each row contains a count of 1. Thus,
The result, above, shows that there was 1 male with red hair, 1 female with blond hair, 2 females with brown hair, and so on. Notice that combinations with zero counts, such as females with red hair, are not displayed.
Another way to generate a table of counts is with the table function. Here is an example of its use and its output:
The result, above, displays a table of all levels of gender crossed with all levels of hair color, explicitly marking combinations that do not occur with counts of zero. This form of output can be useful for human comprehension. But because there are several data values per row, it does not comply with the usual format for data files that are used in this book, which assume a single measurement “instance” per row. The format produced by the aggregate function, in the previous paragraph, is appropriate for the usual data files.
The apply function is handy for collapsing arrays across specified dimensions, and applying a function to the data within the collapsed dimensions. For example, recall from Section 3.4.3 the three-dimensional array named a:
Suppose we want to know the sums of the values within columns and layers, collapsed across rows. This means we want to retain the second and third dimensions, while collapsing across the other dimension. The appropriate command is:
Notice that the MARGIN= argument specifies which dimensions to retain, uncollapsed. If the dimensions are named, you can specify the retained dimensions by their names instead of by their indexical number.
The melt command, from the reshape2 package, is very useful for rearranging data so that there is one datum per row. To use the package, you must first install it on your computer:
You only need to install the package once, then never again. For any new session of R, however, you must load the package into R's working memory, like this:
At this point, R knows the various commands defined in the reshape2 package. To illustrate the melt command, first recall the array a defined earlier:
Notice that the array is three-dimensional, with a datum in every one of the 24 cells. We would like to rearrange the data so that there is one datum per row, with each row also specifying the levels of the array from which the datum came. This is done by the melt command:
Notice that the values that were in the array a, namely the numbers 1-24, are now arranged one per row, in the column named “value.” The result of melt is a data frame, with the level identifiers being factors. You can read more about the many other capabilities of the reshape2 package in the article by Wickham (2007), and in the reference manual available at http://cran.r-project.org/web/packages/reshape2/.
3.7 Programming in R
Instead of typing all the commands one at a time at the command line, you can type them into a text document and then have R execute the document. The document is called a program or script. We saw an example in the program SimpleGraph.R, back in Section 3.2.
RStudio is a nice environment for developing programs in R. If you have associated filenames that end in “.R” with RStudio, then opening the file will open it in RStudio's editing window. For starting a new program, in RStudio use the pull-down menu items File → New → R Script. A blank editing window will open, where you can type your program.
Some important points for newbie programmers:
• Be sure you save your program in a folder where you can find it again, with a filename that is easy to recognize weeks later.
• Be sure to save the program every time you make a small working change.
• If you are about to make a big change, save the current working version and start the modified version with a new filename. This way, when your modified version doesn’t work, you still have the old working version to fall back on.
• Put lots of explanatory comments in your code, so that you can understand what you were doing when you come back to the program months later. To include a comment in a program, simply type a “#” character, and everything after that character, on the same line, will be ignored by the R interpreter. There is a peril inherent in comments, however: They easily become obsolete and confusing if you change the programming code without also changing the corresponding comment. This is a problem I frequently encounter in my own programming, and I hope that the programs I’ve created for this book do not have too many obsolete comments.
3.7.1 Variable names in R
You should name variables meaningfully, so that the programming commands are easy for a reader to understand. If you name your variables cryptically, you will curse your poor judgment when you return to the program weeks later and you have no idea what your program does.
You can use fairly long, descriptive names. If the names get too long, however, then the program becomes unwieldy to type and read. For example, suppose you want to name the crucial final output of a program. You could name it tempfoo, but that's not very meaningful, and might even lead you to think that the variable is unimportant. Instead, you could name it theCrucialFinalOutputThatWillChangeTheWorldForever, but that would be burdensome to type and read as it gets reused in the program. So, you might best name it something like finalOutput, which is meaningful but not too long.
Computer programmers typically use a naming convention called camelBack notation. This is a way of connecting several words into a contiguous variable name without using spaces between words. For example, suppose you want to name a variable “final output.” You are not allowed to name a variable with a space in it because R (and most other languages) interprets spaces as separators of variables. One way to avoid using spaces is to connect the words with explicit connectors such as an underscore or a dot, like this: final_output or final.output. Many programmers do use and recommend those naming conventions. But the underscore notation can be difficult to read in some displays, and the dot notation is interpreted by some programming languages (including R in some contexts) as referring to subcomponents or classes of structured variables, which can confuse people (or computers) who are familiar with that meaning of a dot. Therefore, the spaces are simply dropped, with successive words capitalized: finalOutput. The initial word is typically not capitalized, but some people have different uses for initial-capitalized variable names. R is case sensitive: the variable myVar is different than the variable myvar!
I will try to use camelBack notation in all the programs in this book. I may occasionally lapse from bactrian beauty, instead slithering into snakeback notation (finaloutput) or gooseneck notation (final_output) or ant notation (final.output). If you see these lower forms, quietly shun them, knowing that when you create your own programs, you will use the more highly evolved dromedary design.
3.7.2 Running a program
Running a program is easy, but exactly how to do it depends on how you are interacting with R.
Important: First set the working directory. Many programs read data from another computer file, or save data to another file. The programs need to know where to find or put those files. The default location might have no relevance to your work and may cause problems. Therefore, you must be sure that R knows what directory (also known as a folder) to use for getting and saving data. This location is called the working directory. Typically you want the working directory to be the folder in which the current program resides. In R's basic command window, set the working directory by selecting menu items File → Change dir. In RStudio, set the working directory by selecting menu items Session → Set Working Directory. (The menu structures occasionally change with updated versions of R and RStudio.) In RStudio, to be sure that the working directory really is set properly, look in the lower-right window and click the Files tab. Then click the circular arrow to the right of the files tab to refresh the file listing. Check that it displays the items in the intended working directory. If it does not show the intended working directory contents, then use the folder navigation links at the top of the list of files to find the desired folder, and click the tab marked “More” to set the working directory.
As has been described previously, the easiest way to have R's working directory set to the folder of your program is by invoking RStudio via the program, instead of by opening RStudio before opening the program. With R and RStudio closed, open the desired .R program file, for example in Windows by double clicking the file icon. If files of type .R are not yet associated with an application, your computer will ask you what application to use to open the file. Select the application RStudio, and select the option that tells the computer always to use RStudio for files of that type. Then RStudio will open with the .R program open in its editing window, and, importantly, with that program's folder as R's working directory.
To run an entire program, we tell R to get its commands from a source other than the interactive command line. We give it the source command at the interactive command prompt, to tell it to accept all the commands from the alternative source. For example, to run the SimpleGraph.R program from Section 3.2, we could just type source(“SimpleGraph.R”). This command will work only if the file SimpleGraph.R is in R's current working directory.
Typically you will be working interactively with a program that is open in an editing window. If you have already associated filenames that have .R extensions with RStudio, then when you open the file it will automatically open in RStudio's editing window. Alternatively, you can navigate to a program from RStudio's menu, using File → Open File. Be sure to set the working directory appropriately (see above).
Once the program is open in an editing window, it is easy to run all or only part of it via menu buttons on the editor. For example, in RStudio, the top right of the editing window has buttons marked “Run” and “Source.” The Run button will run the line on which the cursor is presently positioned, or multiple lines if they are presently selected, and will echo the lines in the command window. The Source button will run the entire program without echoing the lines in the command window.
Notice that the source function is different than the load function. The source function reads the referred-to text file as if it were a script of commands for R to execute. The commands might create new variables, but they are read as commands. The load function, on the other hand, expects to read a file that is in compressed Rdata format (not a text file) and specifies variables and their values.
3.7.3 Programming a function
A function takes input values, called “arguments,” and does something with them. In general, a function in R in defined by code of the form:
The commands inside the curly braces can extend over many lines of code. When the function is called, it takes the values of the arguments in parentheses and uses them in the commands in the braces. You invoke the function by commanding R thus:
As a simple example, consider this definition:
The function is named asqplusb and it takes the value a, squares it, and adds the value b. The function then returns the result as output. Here is an example:
In a function call, explicitly labeled arguments may go in any order:
However, arguments without explicit labels must be in the order used in the definition of the function. Notice that these two function calls give different results:
The function definition gave argument b a default value of 1 by specifying b=1 in the list of arguments. This means that if the function is called without an explicit value for b provided, the default value will be used. The argument a was not given a default value, however, and must always be specified in a call to the function. Consider these examples:
In the last example above, there was only one unlabeled argument provided in the function call. R assigns that value to the first argument in the definition, regardless of whether the argument was defined with a default value.
3.7.4 Conditions and loops
There are many situations in which we want a command to be executed only under certain conditions. A special case of that is executing a command a certain number of times. The R language can be programmed with conditions and loops to satisfy these needs.
The basic conditional statement uses an if-else structure. This might be best explained by example. Suppose we want to enter a value, x, and have R reply with “small” if x ≤ 3 and with “big” if x > 3. We can type
If we tell R that x=5before executing the lines above, then the word “big” appears in the command window as a reply from R.
Notice the arrangement of curly braces across lines in the if-else structure above. In particular, the line containing “else” begins with a closing curly brace, which tells R that the “else” clause is continuing the preceding “if.” A line that begins with “else” causes an error:
There are a variety of ways to have R execute a set of commands repeatedly. The most basic is the for loop. We specify a vector of values that R steps through, executing a set of commands for each value of the vector. Here is an example:
Notice that the index countDown took on each of the values in the vector 5:1, and the command show(countDown) was executed. The entries in the vector do not need to be numeric. For example, they can be strings:
There will be many examples of conditions and loops in the programs used later in the book. R has other ways for constructing loops and conditions (such as while), and ways for breaking out of loops (such as break). Type ?”Control” to read about them.
3.7.5 Measuring processing time
It can be useful to know how long a process is taking in R, either to anticipate when a very long process will be completed, or to assess where a program is being inefficient. A simple way to measure processing time is with the proc.time function, which returns the current computer-system time. To measure the duration of a process, use proc.time at the beginning and end of the process, and compute the difference between the times. Here is an example:
In other words, it took R 3.17 seconds (on my computer) to compute the logarithm of every integer from 1 to 1,000,000 and store the results in a vector. Instead of using a loop, let's do the same computation in vectorized form and see how long it takes:
You can see that the vector operation took only about 3% of the time needed for the loop! In general, for loops are slow relative to vectorized operations.
3.7.6 Debugging
This book includes dozens of programs that can be used without modification for real research; all you need to do is load your data. An important goal of the book is also for you to be able to modify the programs for application to different data structures and models. For this purpose, you will need to be able to debug programs as you modify them. To “debug” means to find, diagnose, and correct errors in programs.
To avoid producing errors in the first place, here are a few hints:
• When creating a new program, always start with a new file name. Do not overwrite existing, working programs with a new program of the same name.
• When creating for loops, beware of naming the for-loop index as merely a single letter such as “i”or “j”. Such single letters are extremely easy to confuse and mistype and misperceive! Instead, name the indices meaningfully and distinctly for each index. For example, name a row index something like rowIdx instead of just i, and name a column index something like colIdx instead of just j.
• Use explicit parentheses to be sure that operators are applied in the intended order. In particular, beware of operator precedence when using the colon operator to define index limits in for loops. For example, suppose there are N components of vector theVector and you want to display all but the last two components. This command, for ( vIdx in 1:N-2 ) { show(theVector[vIdx]) }, will not do what you want! There should be parentheses around N-2.
• Use visual white space to make your code easily readable by real human beings. Some R programmers like to suppress spaces because it makes the code more compact. But in my experience, space-free code can be extremely difficult to read and can be much more prone to errors.
• Use indenting to meaningfully group sections of the program. In particular, use RStudio's automatic indenting and parenthesis matching to help keep track of embedded loops and functions. In RStudio, select a section of a program or the entire program and then press ctrl-I (or menu Code → Reindent Lines) to see embedded sections properly indented.
A list of common mistakes in R could go on and on. For example, it is easy to mistakenly use a single equal sign, =, when intending to test for equality, which should use a double equal sign, == (or the all.equal function instead). If you want to check for the maximum or minimum across components of a set of vectors, it is easy to mistakenly use the min function instead of the pmin function. The pmin function has not been discussed above, but it will be described in Section 5.5. If you use the attach function successively on two different data frames that happen to have some shared column names, then the later attached variables will mask the earlier variables. (The attach function has not been previously discussed; you know how to get help.) It is natural to encounter many mistakes as you advance through programming in any language. But do not be deterred because, as an anonymous sage once remarked, “If we learn from our mistakes, shouldn’t we try to make as many mistakes as possible?”
When you encounter an error, here are some hints regarding how to diagnose and repair the problem.
• The error messages displayed by R can sometimes be cryptic but are usually very helpful to isolate where the error occurred and what caused it. When an error message appears, don’t just ignore what it says. Be sure to actually read the error message and see if it makes sense.
• Isolate the first point in the code that causes an error. Run the code sequentially a line at a time, or in small blocks, until the first error is encountered. Fix this error, and it might cure subsequent problems too.
• When you have found that a particular line of code causes an error, and that line involves a complex nesting of embedded functions, check the nested functions from the inside out. Do this by selecting, with the cursor, the inner-most variable or function and running it. Then work outward. An example of doing this is presented in Section 4.5.
• If you have defined a new function that had been working but mysteriously has stopped working, be sure that it is not relying on a variable that is defined outside the function. For examples, suppose that you specify N=30 at the command line without putting it into the program. Then, in the program, you define a function, addN = function( x ) { x+N }. The function will work without complaint until you start a new R session that does not have N defined. In general, be wary of using variables inside a function without explicitly putting them as arguments in the function definition.
When you are dealing with complex programs that involve functions calling functions calling functions, the simple suggestions above can be inadequate. R has several advanced facilities devoted to interactive debugging. These include the functions debug, browser, and traceback. A thorough explanation of these functions could easily fill a large chapter by itself, and the required examples would be more complex than we can tackle after only this short introduction to R. Therefore I invite you to investigate the functions in more detail when you discover need for them. RStudio also has editing facilities for advanced debugging. You can investigate them at the RStudio web page, http://www.rstudio.com/ide/docs/debugging/overview.
3.8 Graphical plots: Opening and saving
Creating and saving graphs in R is easy. Unfortunately, because graphical devices are different on different computer operating systems, the R commands for opening display windows and saving graphs can be different on different systems. I have created simple functions for opening graphical windows and saving their contents that can be used identically on Windows, Macintosh, and Linux operating systems. The functions are openGraph and saveGraph, and they are defined in the file DBDA2E-utilities.R. Here is an example of their use:
Notice that the first line above sourced the program DBDA2E-utilities.R, which holds the definitions of the functions. If that program is not in the current working directory, then R will not know where to find the program and R will return an error. Therefore, be sure to have the program in your current working directory. The saveGraph command saves the graph in the current working directory, so make sure that the working directory is set appropriately.
The openGraph command takes width and height arguments, as shown in the example above. It defaults to a 7 × 7 window if no width and height are specified. The saveGraph command takes file and type arguments. The file should specify the root part of the filename, because the saveGraph function will append the format type as an extension to the filename. The saveGraph function allows the following formats: pdf, eps (encapsulated postscript), jpg, jpeg (same as jpg), and png. You can use multiple saveGraph commands successively to save the same graph in different formats.
Chapter 4.5 (p. 93) provides an example of using various other arguments in the plot command. The example in that section also shows some ways to superimpose lines and annotations on a plot.
3.9 Conclusion
This chapter has surveyed only the most basic ideas and capabilities of R. It is worth repeating here that an excellent summary of basic R functions is available at http://cran.r-project.org/doc/contrib/Short-refcard.pdf. R has many elaborate packages that have been developed for different applications. I would list various web resources here, but aside from the main sites for R and RStudio, other sites are continuously evolving and changing. Therefore, search the web for the latest packages and documentation. There are also numerous books about R and specific applications of R.
3.10 Exercises
Look for more exercises at https://sites.google.com/site/doingbayesiandataanalysis/