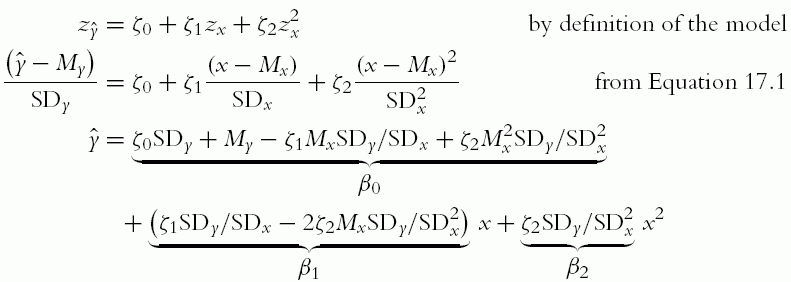Metric Predicted Variable with One Metric Predictor
The agri-bank's threatnin' to revoke my lease
If my field's production don't rapid increase.
Oh Lord how I wish I could divine the trend,
Will my furrows deepen? and Will my line end?1
In this chapter, we consider situations such as predicting a person's weight from their height, or predicting their blood pressure from their weight, or predicting their income from years of education. In these situations, the predicted variable is metric, and the single predictor is also metric.
We will initially describe the relationship between the predicted variable, y, and predictor, x, with a simple linear model and normally distributed residual randomness in y. This model is often referred to as “simple linear regression.” We will generalize the model in three ways. First, we will give it a noise distribution that accommodates outliers, which is to say that we will replace the normal distribution with a t distribution as we did in the previous chapter. The model will be implemented in both JAGS and Stan. Next, we will consider differently shaped relations between the predictor and the predicted, such as quadratic trend. Finally, we will consider hierarchical models of situations in which every individual has data that can be described by an individual trend, and we also want to estimate group-level typical trends across individuals.
In the context of the generalized linear model (GLM) introduced in Chapter 15, this chapter's situation involves a linear function of a single metric predictor, as indicated in the third column of Table 15.1 (p. 434), with a link function that is the identity along with a normal distribution for describing noise in the data, as indicated in the first row of Table 15.2 (p. 443). For a reminder of how this chapter's combination of predicted and predictor variables relates to other combinations, see Table 15.3 (p. 444).
17.1 Simple linear regression
Figure 17.1 shows examples of simulated data generated from the model for simple linear regression. First a value of x is arbitrarily generated. At that value of x, the mean predicted value of y is computed as μ = β0 + β1x. Then, a random value for the datum y is generated from a normal distribution centered at μ with standard deviation σ. For a review of how to interpret the slope, β1, and intercept, β0, see Figure 15.1 (p. 425).

Note that the model only specifies the dependency of y on x. The model does not say anything about what generates x, and there is no probability distribution assumed for describing x. The x values in the left panel of Figure 17.1 were sampled randomly from a uniform distribution, merely for purposes of illustration, whereas the x values in the right panel of Figure 17.1 were sampled randomly from a bimodal distribution. Both panels show data from the same model of the dependency of y on x.
It is important to emphasize that the model assumes homogeneity of variance: At every value of x, the variance of y is the same. This homogeneity of variance is easy to see in the left panel of Figure 17.1: The smattering of data points in the vertical, y, direction appears visually to be the same at all values of x. But that is only because the x values are uniformly distributed. Homogeneity of variance is less easy to identify visually when the x values are not uniformly distributed. For example, the right panel of Figure 17.1 displays data that may appear to violate homogeneity of variance, because the apparent vertical spread of the data seems to be larger at x = 2.5 than at x = 7.5 (for example). Despite this deceiving appearance, the data do respect homogeneity of variance. The reason for the apparent violation is that for regions in which x is sparse, there is less opportunity for the sampled y values to come from the tails of the noise distribution. In regions where x is dense, there are many opportunities for y to come from the tails.
In applications, the x and y values are provided by some real-world process. In the real-world process, there might or might not be any direct causal connection between x and y. It might be that x causes y, or y causes x, or some third factor causes both x and y, or x and y have no causal connection, or some combination of those from multiple types of causes. The simple linear model makes no claims about causal connections between x and y. The simple linear model merely describes a tendency for y values to be linearly related to x values, hence “predictable” from the x values. When describing data with this model, we are starting with a scatter plot of points generated by an unknown process in the real world, and estimating parameter values that would produce a smattering of points that might mimic the real data. Even if the descriptive model mimics the data well (and it might not), the mathematical “process” in the model may have little if anything to do with the real-world process that created the data. Nevertheless, the parameters in the descriptive model are meaningful because they describe tendencies in the data.
17.2 Robust linear regression
There is no requirement to use a normal distribution for the noise distribution. The normal distribution is traditional because of its relative simplicity in mathematical derivations. But real data may have outliers, and the use of (optionally) heavy-tailed noise distributions is straight forward in contemporary Bayesian software. Section 16.2 (p. 458) explained the t distribution and its usefulness for describing data that might have outliers.
Figure 17.2 illustrates the hierarchical dependencies in the model. At the bottom of the diagram, the datum yi is a t-distributed random value around the central tendency μi = β0 + β1xi. The rest of the diagram illustrates the prior distribution on the four parameters. The scale parameter σ is given a noncommittal uniform prior, and the normality parameter v is given a broad exponential prior, as they were in the previous chapter. The intercept and slope are given broad normal priors that are vague on the scale of the data. Figure 17.2 is just like the diagram for robust estimation of the central tendency of groups, shown in Figure 16.11 on p. 468, except here the two parameters β0 and β1 are shown with separate normal priors, whereas in Figure 16.11, the two parameters μ1 and μ2 had their normal priors superimposed.

As an example, suppose we have measurements of height, in inches, and weight, in pounds, for some randomly selected people. Figures 17.3 and 17.4 show two examples, for data sets of N = 30 and N = 300. At this time, focus your attention only on the scatter of data points in the top panels of each figure. The superimposed lines and curves will be explained later. You can see that the axes of the top panels are labeled height and weight, so each point represents a person with a particular combination of height and weight. The data were generated from a program I created (HtWtDataGenerator.![]() ) that uses realistic population parameters (Brainard & Burmaster, 1992). The simulated data are actually based on three bivariate Gaussian clusters, not a single linear dependency. But we can still attempt to describe the data with linear regression. The scatter plots in Figures 17.3 and 17.4 do suggest that as height increases, weight also tends to increase.
) that uses realistic population parameters (Brainard & Burmaster, 1992). The simulated data are actually based on three bivariate Gaussian clusters, not a single linear dependency. But we can still attempt to describe the data with linear regression. The scatter plots in Figures 17.3 and 17.4 do suggest that as height increases, weight also tends to increase.


Our goal is to determine what combinations of β0, β1, σ , and v are credible, given the data. The answer comes from Bayes' rule:
Fortunately, we do not have to worry much about analytical derivations because we can let JAGS or Stan generate a high resolution picture of the posterior distribution. Our job, therefore, is to specify sensible priors and to make sure that the MCMC process generates a trustworthy posterior sample that is converged and well mixed.
17.2.1 Robust linear regression in JAGS
As has been emphasized (e.g., at the end of Section 8.2), hierarchical dependency diagrams like Figure 17.2 are useful not only for conceptualizing the structure of a model but also for programming it. Every arrow in Figure 17.2 has a corresponding line of code in JAGS or Stan. The following sections describe implementations of the model. The core of the model specification for JAGS is virtually an arrow-to-line transliteration of the diagram. The seven arrows in Figure 17.2 have seven corresponding lines of code:
Some of the parameter names in the code above are preceded by the letter “z” because the data will be standardized before being sent to the model, as will be described later, and standardized data often are referred to by the letter “z.”
One of the nice features of JAGS (and Stan) is that arguments in distributions are functionally evaluated, so we can put a statement with operations in the place of an argument instead of only a variable name. For example, in the dt distribution earlier, the precision argument has the expression 1/zsigmaˆ2 instead of just a variable name. We could do the same thing for dt's other arguments. In particular, instead of assigning a value to mu[i] on a separate line, we could simply insert zbeta0 + zbeta1 * zx[i] in place of mu[i] in the mean argument of dt. Thus, assignment arrows in diagrams like Figure 17.2 do not necessarily need a separate line in the JAGS code. However, when the expression is put directly into an argument, JAGS cannot record the value of the expression throughout the chain. Thus, in the form shown earlier, mu[i] is available for recording along with the parameter values at every step. We are not interested in mu[i] at every step; therefore, the revised code (exhibited later) will put its expression directly into the mean argument of dt. On the other hand, the expression for nu will remain on a separate line (as shown above) because we do want an explicit record of its value.
17.2.1.1 Standardizing the data for MCMC sampling
In principle, we could run the JAGS code, shown earlier, on the “raw” data. In practice, however, the attempt often fails. There's nothing wrong with the mathematics or logic; the problem is that credible values of the slope and intercept parameters tend to be strongly correlated, and this narrow diagonal posterior distribution is difficult for some sampling algorithms to explore, resulting in extreme inefficiency in the chains.
You can get an intuition for why this happens by considering the data in the upper panel of Figure 17.3. The superimposed lines are a smattering of credible regression lines from the posterior MCMC chain. All the regression lines must go through the midst of the data, but there is uncertainty in the slope and intercept. As you can see, if the slope is steep, then the y intercept (i.e., the y value of the regression line when x = 0) is low, at a large negative value. But if the slope is not so steep, then the y intercept is not so low. The credible slopes and intercepts trade off. The middle-right panel of Figure 17.3 shows the posterior distribution of the jointly credible values of the slope β1 and the intercept β0. You can see that the correlation of the credible values of the slope and intercept is extremely strong.
MCMC sampling from such a tightly correlated distribution can be difficult. Gibbs sampling gets stuck because it keeps “bumping into the walls” of the distribution. Recall how Gibbs sampling works: One parameter is changed at a time. Suppose we want to change β0 while β1 is constant. There is only a very small range of credible values for β0 when β1 is constant, so β0 does not change much. Then we try changing β1 while β0 is constant. We run into the same problem. Thus, the two parameter values change only very slowly, and the MCMC chain is highly autocorrelated. Other sampling algorithms can suffer the same problem if they are not able to move quickly along the long axis of the diagonal distribution. There is nothing wrong with this behavior in principle, but in practice, it requires us to wait too long before getting a suitably representative sample from the posterior distribution.
There are (at least) two ways to make the sampling more efficient. One way is to change the sampling algorithm itself. For example, instead of using Gibbs sampling (which is one of the main methods in JAGS), we could try Hamiltonian Monte Carlo (HMC) in Stan. We will explore this option in a later section. A second way is to transform the data so that credible regression lines do not suffer such strong correlation between their slopes and intercepts. It is this option we explore presently.
Look again at the data with the superimposed regression lines in Figure 17.3. The problem arises because the arbitrary position of zero on the x axis is far away from the data. If we simply slid the axis so that zero fell under the mean of the data, then the regression lines could tilt up or down without any big changes on the y intercept. Sliding the axis so that zero falls under the mean is equivalent to subtracting the mean of the x values from each x value. Thus, we can transform the x values by “mean centering” them. This alone would solve the parameter-correlation problem.
But if we're going to bother to mean center the x values, we might as well completely standardize the data. This will allow us to set vague priors for the standardized data that will be valid no matter the scale of the raw data. Standardizing simply means re-scaling the data relative to their mean and standard deviation:
where Mx is the mean of the x values and SDx is the standard deviation of the x values. It is easy to prove, using simple algebra, that the mean of the resulting zx values is zero, and the standard deviation of the resulting zx values is one, for any initial data set. Thus, standardized values are not only mean centered, but are also stretched or shrunken so that their standard deviation is one.
We will send the standardized data to the JAGS model, and JAGS will return credible parameter values for the standardized data. We then need to convert the parameter values back to the original raw scales. Denote the intercept and slope for standardized data as ζ0 and ζ1 (Greek letter “zeta”), and denote the predicted value of y as ŷ. Then:
Thus, for every combination of ζ0, ζ1 values, there is a corresponding combination of β0, β1 values specified by Equation 17.2. To get from the σ value of the standardized scale to the σ value on the original scale, we simply multiply by SDy. The normality parameter remains unchanged because it refers to the relative shape of the distribution and is not affected by the scale parameter.2
There is one more choice to make before implementing this method: Should we use ![]() to standardize the data, then run JAGS on the standardized data, and then in
to standardize the data, then run JAGS on the standardized data, and then in ![]() transform the parameters back to the original scale? Or, should we send the original data to JAGS, and do the standardization and transformation back to original scale, all in JAGS? Either method can work. We will do the transformation in JAGS. The reason is that this method keeps an integrated record of the standardized and original-scale parameters in JAGS' coda format, so convergence diagnostics can be easily run. This method also gives me the opportunity to show another aspect of JAGS programming that we have not previously encountered, namely, the data block.
transform the parameters back to the original scale? Or, should we send the original data to JAGS, and do the standardization and transformation back to original scale, all in JAGS? Either method can work. We will do the transformation in JAGS. The reason is that this method keeps an integrated record of the standardized and original-scale parameters in JAGS' coda format, so convergence diagnostics can be easily run. This method also gives me the opportunity to show another aspect of JAGS programming that we have not previously encountered, namely, the data block.
The data are denoted as vectors x and y on their original scale. No other information is sent to JAGS. In the JAGS model specification, we first standardize the data by using a data block, like this:
All the values computed in the data block can be used in the model block. In particular, the model is specified in terms of the standardized data, and the parameters are preceded by a letter z to indicate that they are for the standardized data. Because the data are standardized, we know that the credible slope cannot greatly exceed the range ±1, so a standard deviation of 10 on the slope makes it extremely flat relative to its possible credible values. Similar considerations apply to the intercept and noise, such that the following model block has extremely vague priors no matter what the original scale of the data:
Notice at the end of the model block, above, the standardized parameters are transformed to the original scale, using a direct implementation of the correspondences in Equation 17.2.
A high-level script for using the JAGS model above is named Jags-Ymet-Xmet-Mrobust-Example.R, with functions themselves defined in Jags-Ymet-Xmet-Mrobust.R.![]() unning the script produces the graphs shown in Figures 17.3 and 17.4. Before discussing the results, let's explore how to implement the model in Stan.
unning the script produces the graphs shown in Figures 17.3 and 17.4. Before discussing the results, let's explore how to implement the model in Stan.
17.2.2 Robust linear regression in Stan
![]() ecall from Section 14.1 (p. 400) that Stan uses Hamiltonian dynamics to find proposed positions in parameter space. The trajectories use the gradient of the posterior distribution to move large distances even in narrow distributions. Thus, HMC by itself, without data standardization, should be able to efficiently generate a representative sample from the posterior distribution.
ecall from Section 14.1 (p. 400) that Stan uses Hamiltonian dynamics to find proposed positions in parameter space. The trajectories use the gradient of the posterior distribution to move large distances even in narrow distributions. Thus, HMC by itself, without data standardization, should be able to efficiently generate a representative sample from the posterior distribution.
17.2.2.1 Constants for vague priors
The only new aspect of the Stan implementation is the setting of the priors. Because the data will not be standardized, we cannot use the constants from the priors in the JAGS model. Instead, we need the constants in the priors to be vague on whatever scale the data happen to fall on. We could query the user to supply a prior, but instead we will let the data themselves suggest what is typical, and we will set the priors to be extremely broad relative to the data (just as we did in the previous chapter).
A regression slope can take on a maximum value of SDy/SDx for data that are perfectly correlated. Therefore, the prior on the slope will be given a standard deviation that is large compared to that maximum. The biggest that an intercept could be, for data that are perfectly correlated, is MxSDy/SDx. Therefore, the prior on the intercept will have a standard deviation that is large compared to that maximum. The following model specification for Stan implements these ideas:
A script for the Stan model above is named Stan-Ymet-Xmet-Mrobust-Example. R, with functions defined in Stan-Ymet-Xmet-Mrobust.R.![]() unning it produces graphs just like those shown in Figures 17.3 and 17.4. Before discussing the results, let's briefly compare the performance of Stan and JAGS.
unning it produces graphs just like those shown in Figures 17.3 and 17.4. Before discussing the results, let's briefly compare the performance of Stan and JAGS.
17.2.3 Stan or JAGS?
The Stan and JAGS implementations were both run on the data in Figure 17.4, and they produced very similar results in terms of the details of the posterior distribution. For a comparison, both implementations were run with 500 adaptation steps, 1000 burn-in steps, and 20,000 saved steps with no thinning, using four chains. On my modest desktop computer, a run in JAGS took about 180 seconds, while a run in Stan (including compilation) took about 485 seconds (2.7 times as long). The ESS in JAGS was quite different for different parameters, being about 5000 for v and σ but about 16,000 for β0 and β1. The effective sample size (ESS) in Stan was more consistent across parameters but not as high as JAGS for some, being about 8000 for v and σ but only about 7000 for β0 and β1.
Thus, Stan does indeed go a long way toward solving the correlated-parameter problem for which JAGS needed the intervention of standardizing the data. Stan also samples the normality and scale parameters with less autocorrelation than JAGS, as we saw previously in Figure 16.10 (p. 467). HMC deftly navigates through posterior distributions where other samplers may have troubles. But Stan took longer in real time and yielded a smaller ESS for the main parameters of interest, namely the slope and intercept. Stan's efficiency with this model might be improved if we used standardized data as we did for JAGS, but a main point of this demonstration was to show that HMC is able to navigate difficult posterior distributions. Stan's efficiency (and JAGS') might also be improved if the t distribution were reparameterized, as explained in the Stan Reference Manual. Although subsequent programs in this section focus on JAGS, keep in mind that it is straight forward to translate the programs into Stan.
17.2.4 Interpreting the posterior distribution
Now that we are through all the implementation details, we will discuss the results of the analysis shown in Figures 17.3 and 17.4. Recall that both figures show results from the same data generator, but Figure 17.3 has only 30 data points while Figure 17.4 has 300. By comparing the marginal distributions of the posterior across the two figures, you can see that the modal estimates of the slope, intercept, and scale are about the same, but the certainty of the estimate is much tighter for 300 data points than for 30 data points. For example, with N = 30, the slope has modal estimate of about 4.5 (pounds per inch), with a 95% HDI that extends from about 2.0 to 7.0. When N = 300, again the slope has modal estimate of about 4.5, but the 95% HDI is narrower, extending from about 3.6 to 5.2.
It is interesting to note that the normality parameter does not have the same estimate for the two data sets. As the data set gets bigger, the estimate of the normality parameter gradually gets smaller, overcoming the prior which remains fairly dominant with N = 30. (![]() ecall the discussion of the exponential prior on v in Section 16.2.1, p. 462.) With larger data sets, there is more opportunity for outliers to appear, which are inconsistent with large values of v. (Of course, if the data are genuinely normally distributed, then larger data sets just reinforce large values of the normality parameter.)
ecall the discussion of the exponential prior on v in Section 16.2.1, p. 462.) With larger data sets, there is more opportunity for outliers to appear, which are inconsistent with large values of v. (Of course, if the data are genuinely normally distributed, then larger data sets just reinforce large values of the normality parameter.)
In some applications, there is interest in extrapolating or interpolating trends at x values sparsely represented in the current data. For instance, we might want to predict the weight of a person who is 50 inches tall. A feature of Bayesian analysis is that we get an entire distribution of credible predicted values, not only a point estimate. To get the distribution of predicted values, we step through the MCMC chain, and at each step, we randomly generate simulated data from the model using the parameter values at that step. You can think of this by looking at the vertically plotted noise distributions in Figure 17.4. For any given value of height, the noise distributions show the credible predicted values of weight. By simulating data from all the steps in the chain, we integrate over all those credible noise distributions. An interesting aspect of the predictive distributions is that they are naturally wider, that is, less certain, the farther away we probe from the actual data. This can be seen most easily in Figure 17.3. As we probe farther from the data, the spread between credible regression lines gets larger, and thus the predicted value gets less certain.
The upper panels of Figures 17.3 and 17.4 show the data with a smattering of credible regression lines and t distributions superimposed. This provides a visual posterior predictive check, so we can qualitatively sense whether there are systematic deviations of the data from the form implied by the model. For example, we might look for nonlinear trends in the data or asymmetry in the distribution of weights. The data suggest that there might be some positive skew in the distribution of weights relative to the regression lines. If this possibility had great theoretical importance or were strong enough to cast doubt on the interpretation of the parameters, then we would want to formulate a model that incorporated skew in the noise distribution. For more information about posterior predictive checks, see Kruschke (2013b). Curiously, in the present application, the robust linear-regression model does a pretty good job of mimicking the data, even though the data were actually generated by three bivariate normal distributions. ![]() emember, though, that the model only mimics the dependency of y on x, and the model does not describe the distribution of x.
emember, though, that the model only mimics the dependency of y on x, and the model does not describe the distribution of x.
17.3 Hierarchical regression on individuals within groups
In the previous applications, the jth individual contributed a single xj, yj pair. But suppose instead that every individual, j, contributes multiple observations of xi|j, yi|j pairs. (The subscript notation i|j means the ith observation within the jth individual.) With these data, we can estimate a regression curve for every individual. If we also assume that the individuals are mutually representative of a common group, then we can estimate group-level parameters too.
One example of this scenario comes from Walker, Gustafson, and Frimer (2007), who measured reading-ability scores of children across several years. Thus, each child contributed several age and reading-score pairs. A regression line can describe each child's reading ability through time, and higher-level distributions can describe the credible intercepts and slopes for the group as a whole. Estimates of each individual are mutually informed by data from all other individuals, by virtue of being linked indirectly through the higher-level distribution. In this case, we might be interested in assessing individual reading ability and rate of improvement, but we might also or instead be interested in estimating the group-level ability and rate of improvement.
As another example, suppose we are interested in how family income varies with the size of the family, for different geographical regions. The U.S. Census Bureau has published data that show the median family income as a function of the number of persons in a family, for all 50 states, the District of Columbia, and Puerto Rico. In this case, each territory is a “subject” and contributes several pairs of data: Median income for family size 2, median income for family size 3, and so on, up to family size 6 or 7. If we had the actual income for each family, we could use those data in the analysis instead of the median. In this application, we might be interested in estimates for each individual region, or in the overall estimate. The estimated trend in each region is informed by the data from all the other regions indirectly through the higher-level distribution that is informed by all the regions.
Figure 17.5 shows some fictitious data for a worked example. The scales for x and y are roughly comparable to the height and weight example from the previous section, but the data are not intended to be realistic in this case. In these data, each subject contributes several x, y pairs. You might think of these as growth curves for individual people, but with the caveat that they are not realistic. The top panel of Figure 17.5 shows the data from all the individuals superimposed, with subsets of data from single individuals connected by line segments. The lower panels of Figure 17.5 show the data separated into individuals. Notice that different individuals contribute different numbers of data points, and the x values for different individuals can be completely different.

Our goal is to describe each individual with a linear regression, and simultaneously to estimate the typical slope and intercept of the group overall. A key assumption for our analysis is that each individual is representative of the group. Therefore, every individual informs the estimate of the group slope and intercept, which in turn inform the estimates of all the individual slopes and intercepts. Thereby we get sharing of information across individuals, and shrinkage of individual estimates toward the overarching mode. We will expand our discussion of shrinkage after twisting our minds around the model specification.
17.3.1 The model and implementation in JAGS
Figure 17.6 shows a diagram of the model. It might seem a bit daunting, but if you compare it with Figure 17.2 on p. 480 you will see that it merely adds a level to the basic model we are already very familiar with. The diagram is useful for understanding all the dependencies in the model, and for programming in JAGS (and Stan) because every arrow in the diagram has a corresponding line in the JAGS model specification.
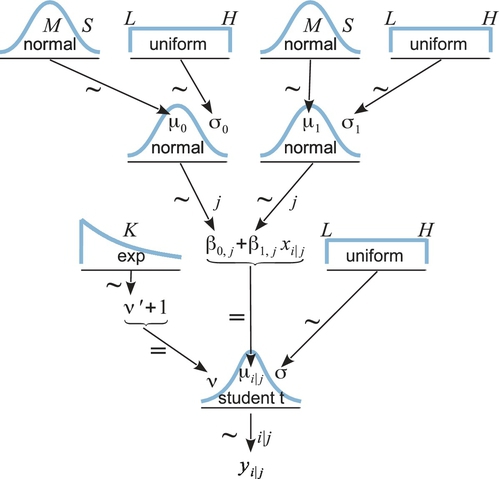
Starting at the bottom of Figure 17.6, we see that the ith datum within the jth subject comes from a t distribution that has a mean of μi|j = β0,j+ β1,jxi|j. Notice that the intercept and slope are subscripted with j, meaning that every subject has its own slope and intercept. Moving up the hierarchical diagram, we see that the slopes across the individuals all come from a higher-level normal distribution that has mean denoted μ1 and standard deviation of σ1, both of which are estimated. In other words, the model assumes that β1,j∼ normal(μ1, σ1), where μ1 describes the typical slope of the individuals and σ1 describes the variability of those individual slopes. If every individual seems to have nearly the same slope, then σ1 is estimated to be small, but if different individuals have very different slopes, then σ1 is estimated to be large. Analogous considerations apply to the intercepts. At the top level of the hierarchy, the group-level parameters are given generic vague priors.
This model assumes that the standard deviation of the noise within each subject is the same for all subjects. In other words, there is a single σ parameter, as shown at the bottom of Figure 17.6. This assumption could be relaxed by providing subjects with distinct noise parameters, all under a higher level distribution. We will see our first example of this approach later, in Figure 19.6 (p. 574).
To understand the JAGS implementation of the model, we must understand the format of the data file. The data format consists of three columns. Each row corresponds to one point of data and specifies the x, y values of the point and the subject who contributed that point. For the ith row of the data file, the x value is x[i], the y value is y[i], and the subject index j is s[i]. Notice that the index i in the JAGS model specification is the row of overall data file, not the ith value within subject j. The model specification assumes that the subject indices are consecutive integers, but the data file can use any sort of unique subject identifiers because the identifiers get converted to consecutive integers inside the program. The subject indices will be used to keep track of individual slopes and intercepts.
The JAGS implementation of this model begins by standardizing the data, exactly like the nonhierarchical model of the previous section:
Then we get to the novel part, where the hierarchical model itself is specified. Notice below that the intercept and slope use nested indexing. Thus, to describe the standardized y value zy[i], the model uses the slope, zbeta1[s[i]], of the subject who contributed that y value. As you read the model block, below, compare each line with the arrows in Figure 17.6:
17.3.2 The posterior distribution: Shrinkage and prediction
A complete high-level script for running the analysis is in Jags-Ymet-XmetSsubj-MrobustHier-Example.R, with the corresponding functions defined in Jags-Ymet-XmetSsubj-MrobustHier.R You will notice that there is nonzero thinning used; this is merely to keep the saved file size relatively small while attaining an ESS of at least 10,000 for relevant parameters. The MCMC file can get large because there are a lot of parameters to store. For example, with 25 subjects, and tracking standardized and original-scale parameters, there are 107 variables recorded from JAGS. The model uses three parallel chains in runjags to save time, but it can still take a few minutes. Occasionally, a run will produce a chain that is well behaved for all parameters except the normality parameter v. This chain nevertheless explores reasonable values of v and is well behaved on other parameters. If this is worrisome, simply run JAGS again until all chains are well behaved (or translate to Stan).
Figure 17.5 shows the data with a smattering of credible regression lines superimposed. The lines on the overall data, in the top panel of Figure 17.5, plot the group-level slope and intercept. Notice that the slopes are clearly positive, even though the collective of data points, disconnected from their individual sources, show no obvious upward trend.
The lower panels of Figure 17.5 show individual data with a smattering of credible regression lines superimposed, using individual-level slopes and intercepts. Because of information sharing across individuals, via the higher-level distribution, there is notable shrinkage of the estimates of the individuals. That is to say, the estimates of the individual slopes are pulled together. This shrinkage is especially clear for the final two subjects, who each have only a single data point. Despite this dearth of data, the estimates of the slope and intercept for these subjects are surprisingly tightly constrained, as shown by the fact that the smattering of credible lines is a fairly tight bundle that looks like a blurry version of the posterior predictive lines in the other subjects. Notice that the estimated intercepts of the two final subjects are pulled toward the data values, so that predictions are different for the different individuals. This is another illustration of shrinkage of estimation in hierarchical models, which was introduced in Section 9.3 (p. 245).
17.4 Quadratic trend and weighted data
The U.S. Census Bureau publishes information from the American Community Survey and Puerto Rico Community Survey, including data about family income and family size.3 Suppose we are interested in the median family income for different family sizes, in each of the 50 states, Puerto Rico, and the District of Columbia. The data are displayed in Figure 17.7. You can see that the incomes appear to have a nonlinear, inverted-U trend as family size increases. Indeed, if you run the data through the linear-regression model of the previous section, you will notice obvious systematic deviations away from the posterior predicted lines.

Because a linear trend appears to be an inadequate description of the data, we will extend the model to include a quadratic trend. This is not the only way to express a curve mathematically, but it is easy and conventional. A quadratic has the form y = b0 + b1x + b2x2. When b2 is zero, the form reduces to a line. Therefore, this extended model can produce any fit that the linear model can. When b2 is positive, a plot of the curve is a parabola that opens upward. When b2 is negative, the curve is a parabola that opens downward. We have no reason to think that the curvature in the family-income data is exactly a parabola, but the quadratic trend might describe the data much better than a line alone. If the posterior distribution on the parameters indicates that credible values of b2 are far less than zero, we have evidence that the linear model is not adequate (because the linear model would have b2 = 0).
We will expand the model to include a quadratic component for every individual (i.e., state) and for the group level. To make the MCMC more efficient, we will standardize the data and then transform the parameters back to the original scale, just as we did for the linear model (see Equation 17.2, p. 485). Because of the quadratic component, the transformation involves a little more algebra:
The correspondences in Equation 17.3 are implemented in JAGS, so JAGS keeps track of the standardized coefficients and the original-scale coefficients.
There is one other modification we will make for the family-income data. The data report the median income at each family size, but the median is based on different numbers of families at each size. Therefore, every median has a different amount of sampling noise. Typically, there are fewer large-sized families than small-sized families, and therefore, the medians for the large-size families are noisier than the medians for the small-size families. The model should take this into account, with parameter estimates being more tightly constrained by the less noisy data points.
Fortunately, the U.S. Census Bureau reported each median with a “margin of error” that reflects the standard error of the data collected for that point. We will use the margin of error to modulate the noise parameter in the model. If the margin of error is high, then the noise parameter should be increased proportionally. If the margin of error is small, then the noise parameter should be decreased proportionally. More formally, instead of using yi∼ normal(μi, σ) with the same σ for every datum, we use yi ∼ normal(μi, wiσ),where wi reflects the relative standard error for the ith datum.4 When wi is small, then μi has relatively little wiggle room around its best fitting value. When wi is large, then μi has can deviate farther from its best fitting value without greatly changing the likelihood of the datum. Overall, then, this formulation finds parameter values that better match the data points with small margin of error than the data points with large margin of error. In the JAGS specification below, the original-scale standard errors are divided by their mean, so that a weight of 1.0 implies the mean standard error.
The quadratic component and weighted noise are implemented in the JAGS model as follows:
In the JAGS specification above, notice that the mean argument of the student t distribution includes the new quadratic component. And notice that the precision argument of the student t distribution multiplies zsigma by the datum-specific noise weight, zw[i]. After the likelihood function, the remaining additions to the model specification merely specify the prior on the quadratic terms and implement the new transformation to the original scale (Equation 17.3). The complete script and functions for the model are in Jags-Ymet-XmetSsubj-MrobustHierQuadWt-Example.R and Jags-Ymet-XmetSsubj-MrobustHierQuadWt.R. The script is set up such that if you have a data file without standard errors for each data point, the weight name is simply omitted (or explicitly set as wName=NULL).
The Stan version of the specification above is provided in Exercise 17.3.
17.4.1 Results and interpretation
As you can see from the superimposed credible regression curves in Figure 17.7, the curvature is quite prominent. In fact, the 95% HDI on the quadratic coefficient (not shown here, but displayed by the script) goes from −2200 to −1700, which is far from zero. Thus, there is no doubt that there is a nonlinear trend in these data.
Interestingly, shrinkage from the hierarchical structure informs the estimates for each state. For example, inspect the subpanel for Hawaii in Figure 17.7. Its credible trend lines are curved downward even though its data in isolation curve upward. Because the vast majority of states show downward curvature in their data, and because Hawaii is assumed to be like the other states, the most credible estimates for Hawaii have a downward curvature. If Hawaii happened to have had a ginormous amount of data, so that its noise weights were extremely small, then its individual credible curves would more closely match its individual trend. In other words, shrinkage from the group level is a compromise with data from the individual.
Care must be taken when interpreting the linear component, that is, the slope. The slope is only meaningful in the context of adding the quadratic component. In the present application, the 95% HDI on the slope goes from 16,600 to 21,800. But this slope, by itself, greatly overshoots the data because the quadratic component subtracts a large value.
The weighting of data by their standard error is revealed by greater uncertainty at large family sizes than small. For example, consider the subpanel for California (or almost every other state) in Figure 17.7. The credible curves have a narrow spread at family size 2 but large spread at family size 7. This is caused in part by the fact that most of the data for large family sizes have large standard errors, and therefore, the parameters have more “slop” or flexibility at the large family sizes. (Another cause of spread in credible curves is variability within and between units.)
Finally, the posterior distribution reveals that these data have outliers within individual states because the normality parameter is estimated to be very small. Almost all of the posterior distribution is below v = 4. This suggests that most of the data points within each individual state fall fairly close to a quadratic-trend line, and the remaining points are accommodated by large tails of the t distribution.
17.4.2 Further extensions
Even this model has many simplifications for the purpose of clarity. One simplification was the use of only linear and quadratic trends. It is easy in Bayesian software to include any sort of non-linear trend, such as higher-order polynomial terms, or sinusoidal trends, or exponential trends or any other mathematically defined trend.
This model assumed a single underlying noise for all individuals. The noise could be modulated by the relative standard error of each datum, but the reference quantity, σ, was the same for all individuals. We do not need to make this assumption. Different individuals might have different inherent amounts of noise in their data. For example, suppose we were measuring a person's blood pressure at various times of day. Some people might have relatively consistent blood pressure while others might have greatly varying blood pressure. When there is sufficient data for each individual, it could be useful to expand the model to include subject-specific standard deviations or “noise” parameters. This lets the less noisy individuals have a stronger influence on the group-level estimate than the noisier individuals. The individual noise parameters could be modeled as coming from a group-level gamma distribution: σi ∼ gamma(r, s), where r and s are estimated, so that the gamma distribution describes the variation of the noise parameter across individuals. We will see our first example of this approach later in Figure 19.6 (p. 574).
Another simplification was assuming that intercepts, slopes, and curvatures are normally distributed across individuals. But it could be that there are outliers: Some individuals might have very unusual intercepts, slopes, or curvatures. Therefore, we could use t distributions at the group level. It is straight forward in Bayesian software to change the distributions from normal to student t. The model thereby becomes robust to outliers at multiple levels. But remember that estimating the normality parameter relies on data in the tails, so if the data set has few individuals and no extreme individuals, there will be nothing gained by using t distributions at the group level.
Another simplification in the model was that it had no explicit parameters to describe covariation of the intercepts, slopes, and curvatures across individuals. For example, rat pups that are born relatively small tend to grow at a smaller rate than rat pups that are born relatively larger: The intercept (weight at birth) and slope (growth rate) naturally covary. In other applications, the correlation could be the opposite, of course. The point is that the present model has no explicit parameter to capture such a correlation. It is straight forward in Bayesian software to use a multivariate normal prior on the intercept, slope, and curvature parameters. The multivariate normal has explicit covariance parameters, which are estimated along with the other parameters. (See, e.g., the “birats” example from WinBUGS Examples Volume 2 at http://www.mrc-bsu.cam.ac.uk/wp-content/uploads/WinBUGS_Vol2.pdf.)
17.5 Procedure and perils for expanding a model
17.5.1 Posterior predictive check
Comparing the data to the posterior predictions of the model is called a posterior predictive check. When there appear to be systematic discrepancies that would be meaningful to address, you should consider expanding or changing the model so it may be a better description of the data.
But what extension or alternative model should you try? There is no uniquely correct answer. The model should be both meaningful and computationally tractable. Meaning can come from mere familiarity in the catalog of functions learned in traditional math courses. This familiarity and simplicity was what motivated the use of a quadratic trend for the family-income data. Alternatively, meaning can come from theories of the application domain. For example, for family income, we might imagine that total income rises as the square root of the number of adults in the family (because each adult has the potential to bring in money to the family) and declines exponentially with the number of children in the family (because each child requires more time away from income generation by the adults). That model is completely fictitious and is mentioned merely for illustration.
The extended model is intended to better describe the data. One way to extend a model is to leave the original model “nested” inside the extended model, such that all the original parameters and mathematical form can be recovered from the extended model by setting the extended parameters to specific constants, such as zero. An example is extending the linear model to a quadratic-trend model; when the curvature is set to zero, the nested linear model is recovered. In the case of nested models, it is easy to check whether the extension fits the data better merely by inspecting the posterior distribution on the new parameters. If the new parameter values are far from the setting that produces the original model, then we know that the new parameters are useful for describing the data.
Instead of adding new parameters to a nested existing model, we might be inspired to try a completely different model form. To assess whether the new model describes the data better than the original model, Bayesian model comparison could be used, in principle. The priors on the parameters within the two models would need to be equivalently informed, as was discussed in Section 10.6.1 (p. 294).
Some people caution that looking at the data to inspire a model form is “double dipping” into the data, in the sense that the data are being used to change the prior distribution on the model space. Of course, we can consider any model space we care to, and the motivation for that model space can evolve through time and come from many different sources, such as different theories and different background literature. If a set of data suggests a novel trend or functional form, presumably the prior probability on that novel trend was small unless the analyst realizes the same trend had occurred unheralded in previous results. The analyst should keep the low prior probability in mind, either formally or informally. Moreover, novel trends are retrospective descriptions of particular data sets, and the trends need to be confirmed by subsequent, independently collected data.
Another approach to a posterior predictive check is to create a posterior predictive sampling distribution of a measure of discrepancy between the predictions and the data. In this approach, a “Bayesian p value” indicates the probability of getting the data's discrepancy, or something more extreme, from the model. If the Bayesian p value is too small, then the model is rejected. From my discussion of p values in Chapter 11, you might imagine that I find this approach to be problematic. And you would be right. I discuss this issue in more detail in an article (Kruschke, 2013b).
17.5.2 Steps to extend a JAGS or Stan model
One of the great benefits of Bayesian software is its tremendous flexibility for specifying models that are theoretically meaningful and appropriate to the data. To take advantage of this flexibility, you will want to be able to modify existing programs. An example of such a modification was presented in this chapter in the extension from the linear-trend model to the quadratic-trend model. The steps in such a modification are routine, as outlined here:
• Carefully specify the model with its new parameters. It can help to sketch a diagram like Figure 17.6 to make sure that you really understand all the parameters and their priors. The diagram can also help coding the model, because there is usually a correspondence of arrows to lines of code. For example when extending a linear model to a quadratic, the diagram gets just one more branch, for the quadratic coefficient, that is structurally identical to the branch for the linear coefficient. The expression for the mean must be extended with the quadratic term: ![]() using notation in the code that is consistent with the already-established code. The group-level distribution over the unit-level quadratic coefficients must also be included. Again, this is easy because the model structure of the quadratic coefficient is exactly analogous to the model structure of the linear coefficient.
using notation in the code that is consistent with the already-established code. The group-level distribution over the unit-level quadratic coefficients must also be included. Again, this is easy because the model structure of the quadratic coefficient is exactly analogous to the model structure of the linear coefficient.
• Be sure all the new parameters have sensible priors. Be sure the priors for the old parameters still make sense in the new model.
• If you are defining your own initial values for the chains, define initial values for all the new parameters, and make sure that the initial values of the old parameters still make sense for the extended model. JAGS will initialize any stochastic variables that are not given explicit initial values. It is often easiest to let JAGS initialize parameters automatically.
• Tell JAGS to track the new parameters. JAGS records only those parameters that you explicitly tell it to track. Stan tracks all nonlocal variables (such as parameters and transformed parameters) by default.
• Modify the summary and graphics output to properly display the extended model. I have found that this step is the most time-consuming and often the most error-prone. Because the graphics are displayed by ![]() , not by JAGS, you must modify all of your
, not by JAGS, you must modify all of your ![]() graphics code to be consistent with the modified model in JAGS. Depending on the role of the parameter in the model and its meaning for interpreting the data, you might want a plot of the parameter's marginal posterior distribution, and perhaps pairs plots of the parameter crossed with other parameters to see if the parameters are correlated in the posterior, and probably specialized displays of the posterior predictive that show trends and predicted spread of data superimposed on the data, and possibly differences between parameters or other comparisons and contrasts, etc.
graphics code to be consistent with the modified model in JAGS. Depending on the role of the parameter in the model and its meaning for interpreting the data, you might want a plot of the parameter's marginal posterior distribution, and perhaps pairs plots of the parameter crossed with other parameters to see if the parameters are correlated in the posterior, and probably specialized displays of the posterior predictive that show trends and predicted spread of data superimposed on the data, and possibly differences between parameters or other comparisons and contrasts, etc.
17.5.3 Perils of adding parameters
When including new parameters, the extended model has new flexibility in fitting the data. There may be a far wider range of credible values for the previous parameters. For example, consider again the data that were introduced for the hierarchical linear model, back in Figure 17.5 (p. 492). We can easily use the hierarchical quadratic-trend model on these data, which yields the posterior credible trends shown in Figure 17.8.5 Notice that there are many positive or negative curvatures that are consistent with the data. The curvature and slope trade-off strongly in their fit to the original-scale data, such that a positive curvature with small slope fits the data as well as a negative curvature with large slope. Thus, the introduction of the curvature parameter makes the slope parameter less certain, even though the posterior distribution of the curvature parameter is centered nearly at zero. Specifically, in the linear-trend fit, wherein the curvature is implicitly forced to be exactly zero, the 95% HDI on the slope goes from +2.2 to +4.1 (not shown here, but produced in the output of the script). In the quadratic-trend fit, the curvature has a mode very near zero with a 95% HDI from about −0.10 to +0.07, but the 95% HDI of the slope goes from about −7.0 to +16.5. In other words, even though the quadratic-trend model indicates that there is little if any quadratic trend in the data, the estimate of the slope is blurred. The magnitude of this apparent blurring of the slope depends strongly on the data scale, however. Using standardized data, the 95% HDI on the slope parameter changes only slightly.

This increase in uncertainty of a parameter estimate is a form of penalizing complexity: For nested models, in which the simpler model is the more complex model with one of the complex model's parameters fixed (e.g., to zero), then the estimates of the nested parameters will tend to be less precise in the more complex model. The degree to which a parameter estimate broadens, when expanding the model, depends on the model structure, the data, and the parameterization. The “blurring” can be especially pronounced if the parameter values can trade-off and fit the data equally well.
A different way of penalizing complexity comes from Bayesian model comparison. The prior on the larger parameter space dilutes the prior probability of any particular combination of parameter values, thereby favoring the simpler model unless the data are much better fit by a combination of parameter values that is not available to the simpler model. It is worth recapitulating that in Bayesian model comparison, the priors for the two models would need to be equivalently informed, as was discussed in Section 10.6.1 (p. 294).
17.6 Exercises
Look for more exercises at https://sites.google.com/site/doingbayesiandataanalysis/