In order for an insurance company to make money, it needs to collect more in yearly premiums than it spends on medical care to its beneficiaries. As a result, insurers invest a great deal of time and money to develop models that accurately forecast medical expenses.
Medical expenses are difficult to estimate because the most costly conditions are rare and seemingly random. Still, some conditions are more prevalent for certain segments of the population. For instance, lung cancer is more likely among smokers than non-smokers, and heart disease may be more likely among the obese.
The goal of this analysis is to use patient data to estimate the average medical care expenses for such population segments. These estimates could be used to create actuarial tables which set the price of yearly premiums higher or lower depending on the expected treatment costs.
For this analysis, we will use a simulated dataset containing medical expenses for patients in the United States. These data were created for this book using demographic statistics from the U.S. Census Bureau, and thus approximately
reflect real-world conditions.
The insurance.csv file includes 1,338 examples of beneficiaries currently enrolled in the insurance plan, with features indicating characteristics of the patient as well as the total medical expenses charged to the plan for the calendar year. The features are:
age: This is an integer indicating the age of the primary beneficiary (excluding those above 64 years, since they are generally covered by the government).sex: This is the policy holder's gender, eithermaleorfemale.bmi: This is the body mass index (BMI), which provides a sense of how over or under-weight a person is relative to their height. BMI is equal to weight (in kilograms) divided by height (in meters) squared. An ideal BMI is within the range of 18.5 to 24.9.children: This is an integer indicating the number of children / dependents covered by the insurance plan.smoker: This isyesornodepending on whether the insured regularly smokes tobacco.region: This is the beneficiary's place of residence in the U.S., divided into four geographic regions:northeast,southeast,southwest, ornorthwest.
It is important to give some thought to how these variables may be related to billed medical expenses. For instance, we might expect that older people and smokers are at higher risk of large medical expenses. Unlike many other machine learning methods, in regression analysis, the relationships among the features are typically specified by the user rather than detected automatically. We'll explore some of these potential relationships in the next section.
As we have done before, we will use the read.csv() function to load the data for analysis. We can safely use stringsAsFactors = TRUE because it is appropriate to convert the three nominal variables to factors:
> insurance <- read.csv("insurance.csv", stringsAsFactors = TRUE)
The str() function confirms that the data are formatted as we had expected:
> str(insurance) 'data.frame': 1338 obs. of 7 variables: $ age : int 19 18 28 33 32 31 46 37 37 60 ... $ sex : Factor w/ 2 levels "female","male": 1 2 2 2 2 1 ... $ bmi : num 27.9 33.8 33 22.7 28.9 ... $ children: int 0 1 3 0 0 0 1 3 2 0 ... $ smoker : Factor w/ 2 levels "no","yes": 2 1 1 1 1 1 ... $ region : Factor w/ 4 levels "northeast","northwest", ... $ charges : num 16885 1726 4449 21984 3867 ...
Since the dependent variable is charges, let's take a look to see how it is distributed:
> summary(insurance$charges) Min. 1st Qu. Median Mean 3rd Qu. Max. 1122 4740 9382 13270 16640 63770
Because the mean value is greater than the median, this implies that the distribution of insurance charges is right-skewed. We can confirm this visually using a histogram:
> hist(insurance$charges)
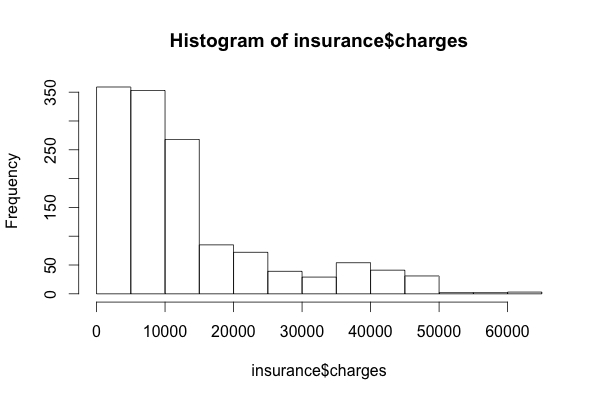
The large majority of individuals in our data have yearly medical expenses between zero and $15,000, although the tail of the distribution extends far past these peaks. Because linear regression assumes a normal distribution for the dependent variable, this distribution is not ideal. In practice, the assumptions of linear regression are often violated. If needed, we may be able to correct this later on.
Another problem at hand is that regression models require that every feature is numeric, yet we have three factor type in our data frame. We will see how R's linear regression function treats our variables shortly.
The sex variable is divided into male and female levels, while smoker is divided into yes and no. From the summary() output, we know that region has four levels, but we need to take a closer look to see how they are distributed.
> table(insurance$region) northeast northwest southeast southwest 324 325 364 325
Here, we see that the data have been divided nearly evenly among four geographic regions.
Before fitting a regression model to data, it can be useful to determine how the independent variables are related to the dependent variable and each other. A correlation matrix provides a quick overview of these relationships. Given a set of variables, it provides a correlation for each pairwise relationship.
To create a correlation matrix for the four numeric variables in the insurance data frame, use the cor() command:
> cor(insurance[c("age", "bmi", "children", "charges")]) age bmi children charges age 1.0000000 0.1092719 0.04246900 0.29900819 bmi 0.1092719 1.0000000 0.01275890 0.19834097 children 0.0424690 0.0127589 1.00000000 0.06799823 charges 0.2990082 0.1983410 0.06799823 1.00000000
At the intersection of each row and column pair, the correlation is listed for the variables indicated by that row and column. The diagonal is always 1 since there is always a perfect correlation between a variable and itself. The values above and below the diagonal are identical since correlations are symmetrical. In other words, cor(x, y) is equal to cor(y, x).
None of the correlations in the matrix are considered strong, but there are some notable associations. For instance, age and bmi appear to have a moderate correlation, meaning that as age increases, so does bmi. There is also a moderate correlation between age and charges, bmi and charges, and children and charges. We'll try to tease out these relationships more clearly when we build our final regression model.
It can also be helpful to visualize the relationships among features, perhaps by using a scatterplot. Although we could create a scatterplot for each possible relationship, doing so for a large number of features might become tedious.
An alternative is to create a scatterplot matrix (sometimes abbreviated as SPLOM), which is simply a collection of scatterplots arranged in a grid. It is used to detect patterns among three or more variables. The scatterplot matrix is not a true multi-dimensional visualization because only two features are examined at a time. Still, it provides a general sense of how the data may be interrelated.
We can use R's graphical capabilities to create a scatterplot matrix for the four numeric features: age, bmi, children, and charges. The pairs() function is provided in a default R installation and provides basic functionality for producing scatterplot matrices. To invoke the function, simply provide it the data frame to present. Here, we'll limit the insurance data frame to the four numeric variables of interest:
> pairs(insurance[c("age", "bmi", "children", "charges")])
This produces the following diagram:

As with the correlation matrix, the intersection of each row and column holds the scatterplot of the variables indicated by the row and column pair. The diagrams above and below the diagonal are transpositions since the x axis and y axis have been swapped.
Do you notice any patterns in these plots? Although some look like random clouds of points, a few seem to display some trends. The relationship between age and charges displays several relatively straight lines, while bmi and charges has two distinct groups of points. It is difficult to detect trends in any of the other plots.
If we add more information to the plot, it can be even more useful. An enhanced scatterplot matrix can be created with the pairs.panels() function in the psych package. If you do not have this package installed, type install packages("psych") to install it on your system then load it using the library(psych) command. Then, we can create a scatterplot matrix as we had done previously:
> pairs.panels(insurance[c("age", "bmi", "children", "charges")])
This produces a slightly more informative scatterplot matrix, as follows:

Above the diagonal, the scatterplots have been replaced with a correlation matrix. On the diagonal, a histogram depicting the distribution of values for each feature is shown. Finally, the scatterplots below the diagonal now are presented with additional visual information.
The oval-shaped object on each scatterplot is a correlation ellipse. It provides a visualization of how strongly correlated the variables are. The dot at the center of the ellipse indicates the point of the mean value for the x axis variable and y axis variable. The correlation between the two variables is indicated by the shape of the ellipse; the more it is stretched, the stronger the correlation. An almost perfectly round oval, as with bmi and children, indicates a very weak correlation (in this case 0.01).
The curve drawn on the scatterplot is called a loess smooth. It indicates the general relationship between the x axis and y axis variables. It is best understood by example. The curve for age and children is an upside-down U, peaking around middle age. This means that the oldest and youngest people in the sample have fewer children than those around middle age. Because this trend is non-linear, this finding could not have been inferred from the correlations alone. On the other hand, the loess smooth for age and bmi is a line sloping gradually up, implying that BMI increases with age, but we had already inferred this from the correlation matrix.
To fit a linear regression model to data with R, the lm() function can be used. This is included in the stats package, which should be included and loaded by default with your R installation. The lm() syntax is as follows:

The following command fits a linear regression model called ins_model, which relates the six independent variables to the total medical charges. The R formula syntax uses the tilde character ~ to describe the model; the dependent variable charges goes to the left of the tilde while the independent variables go to the right, separated by the + sign. There is no need to specify the regression model's intercept term, as it is assumed by default:
> ins_model <- lm(charges ~ age + children + bmi + sex +smoker + region, data = insurance)
Because the . character can be used to specify all features (excluding those already specified in the formula), the following command is equivalent to the preceding command:
> ins_model <- lm(charges ~ ., data = insurance)
After building the model, simply type the name of the model object to see the estimated beta coefficients:
> ins_model3 Call: lm(formula = charges ~ age + children + bmi + sex + smoker + region, data = insurance) Coefficients: (Intercept) age children -11938.5 256.9 475.5 bmi sexmale smokeryes 339.2 -131.3 23848.5 regionnorthwest regionsoutheast regionsouthwest -353.0 -1035.0 -960.1
Understanding the regression coefficients is fairly straightforward. The intercept tells us the value of charges when the independent variables are equal to zero.
Tip
As is the case here, quite often the intercept is difficult to interpret because it is impossible to have values of zero for all features. For example, since no person exists with age zero and BMI zero, the slope has no inherent meaning. For this reason, in practice, the intercept is often ignored.
The estimated beta coefficients indicate the increase in charges for an increase of one in each of the features when the other features are held constant. For instance, for each year that age increases, we would expect $256.90 higher medical expenses on average, assuming everything else is equal. Similarly, each additional child results in an average of $475.50 in additional medical expenses each year, and each unit of BMI increase is associated with an increase of $339.20 in yearly medical costs.
You might notice that although we only specified six features in our model formula, there are eight coefficients reported in addition to the intercept. This happened because the lm() function automatically applied a technique known as dummy coding to each of the factor type variables we included in the model.
Dummy coding allows a nominal feature to be treated as numeric by creating a binary variable for each category of the feature, which is set to 1 if the observation falls into that category or 0 otherwise. For instance, the sex variable has two categories, male and female. This will be split into two binary values, which R names sexmale and sexfemale. For observations where sex = male, then sexmale = 1 and sexfemale = 0; if sex = female, then sexmale = 0 and sexfemale = 1. The same coding applies to variables with three or more categories. The four-category feature region can be split into four variables: regionnorthwest, regionsoutheast, regionsouthwest, and regionnortheast.
When adding a dummy-coded variable to a regression model, one category is always left out to serve as the reference category. The estimates are then interpreted relative to the reference. In our model, R automatically held out the sexfemale, smokerno, and regionnortheast variables, making female non-smokers in the northeast region the reference group. Thus, males have $131.30 less medical costs each year relative to females and smokers cost an average of $23,848.50 more than non-smokers. Additionally, the coefficient for each of the other three regions in the model is negative, which implies that the northeast region tends to have the highest average medical expenses.
The results of the linear regression model make logical sense; old age, smoking, and obesity tend to be linked to additional health issues, while additional family member dependents may result in an increase in physician visits and preventive care such as vaccinations and yearly physical exams. However, we currently have no sense of how well the model is fitting the data. We'll answer this question in the next section.
The parameter estimates we obtained by typing ins_model tell us about how the independent variables are related to the dependent variable, but they tell us nothing about how well the model fits our data. To evaluate the model performance, we can use the summary() command on the stored model:
> summary(ins_model)
This produces the following output:

The summary() output may seem confusing at first, but the basics are easy to pick up. As indicated by the numbered labels in the preceding output, the output provides three key ways to evaluate the performance (that is, fit) of our model:
- The
Residualssection provides summary statistics for the errors in our predictions, some of which are apparently quite substantial. Since a residual is equal to the true value minus the predicted value, the maximum error of29992.8suggests that the model under-predicted expenses by nearly $30,000 for at least one observation. On the other hand, 50 percent of errors fall within the 1Q and 3Q values (the first and third quartile), so the majority of predictions were between $2,850 over the true value and $1,400 under the true value. - The stars (for example,
***)indicate the predictive power of each feature in the model. The significance level (as listed by theSignif.codes in the footer) provides a measure of how likely the true coefficient is zero given the value of the estimate. The presence of three stars indicates a significance level of0, which means that the feature is extremely unlikely to be unrelated to the dependent variable. A common practice is to use a significance level of 0.05 to denote a statistically significant variable. If the model had few features that were statistically significant, it may be cause for concern, since it would indicate that our features are not very predictive of the outcome. Here, our model has several significant variables, and they seem to be related to the outcome in logical ways. - The
Multiple R-squaredvalue (also called the coefficient of determination) provides a measure of how well our model as a whole explains the values of the dependent variable. It is similar to the correlation coefficient in that the closer the value is to 1.0, the better the model perfectly explains the data. Since the R-squared value is0.7494, we know that nearly 75 percent of the variation in the dependent variable is explained by our model. Because models with more features always explain more variation, theAdjusted R-squaredvalue corrects R-squared by penalizing models with a large number of independent variables. It is useful for comparing the performance of models with different numbers of explanatory variables.
Given the preceding three performance indicators, our model is performing fairly well. It is not uncommon for regression models of real-world data to have fairly low R-squared values; a value of 0.75 is actually quite good. The size of some of the errors is a bit concerning, but not surprising given the nature of medical expense data. However, as shown in the next section, we may be able to improve the model's performance by specifying the model in a slightly different way.
As mentioned previously, a key difference between regression modeling and other machine learning approaches is that regression typically leaves feature selection and model specification to the user. Consequently, if we have subject matter knowledge about how a feature is related to the outcome, we can use this information to inform the model specification and potentially improve the model's performance.
In linear regression, the relationship between an independent variable and the dependent variable is assumed to be linear, yet this may not necessarily be true. For example, the effect of age on medical expenditures may not be constant throughout all age values; the treatment may become disproportionately expensive for the oldest populations.
If you recall, a typical regression equation follows a form similar to this:

To account for a non-linear relationship, we can add a higher order term to the regression model, treating the model as a polynomial. In effect, we will be modeling a relationship like this:

The difference between these two models is that a separate beta will be estimated, which is intended to capture the effect of the x-squared term. This allows the impact of age to be measured as a function of age squared.
To add the non-linear age to the model, we simply need to create a new variable:
> insurance$age2 <- insurance$age^2
Then, when we produce our improved model, we'll add both age and age2 to the lm() formula, for example, charges ~ age + age2.
Suppose we have a hunch that the effect of a feature is not cumulative, but rather it has an effect only once a specific threshold has been reached. For instance, BMI may have zero impact on medical expenditures for individuals in the normal weight range, but it may be strongly related to higher costs for the obese (that is, BMI of 30 or above).
We can model this relationship by creating a binary indicator variable that is 1 if the BMI is at least 30 and 0 otherwise. The estimated beta for this binary feature would then indicate the average net impact on medical expenses for individuals with BMI of 30 or above, relative to those with BMI less than 30.
To create the feature, we can use the ifelse() function, which for each element in a vector tests a specified condition and returns a value depending on whether the condition is true or false. For BMI greater than or equal to 30, we will return 1, otherwise 0:
> insurance$bmi30 <- ifelse(insurance$bmi >= 30, 1, 0)
We can then include the bmi30 variable in our improved model, either replacing the original bmi variable or in addition, depending on whether or not we think the effect of obesity occurs in addition to a separate BMI effect. Without good reason to do otherwise, we'll include both in our final model.
So far, we have only considered each feature's individual contribution to the outcome. What if certain features have a combined impact on the dependent variable? For instance, smoking and obesity may have harmful effects separately, but it is reasonable to assume that their combined effect may be worse than the sum of each one alone.
When two features have a combined effect, this is known as an interaction. If we suspect that two variables interact, we can test this hypothesis by adding their interaction to the model. Interaction effects can be specified using the R formula syntax. To interact the obesity indicator (bmi30) with the smoking indicator (smoker), we would write a formula in the form charges ~ bmi30*smoker
The * operator is shorthand that instructs R to model charges ~ bmi30 + smokeryes + bmi30:smokeryes
The : (colon) operator in the expanded form indicates that bmi30:smokeryes is the interaction between the two variables. Note that the expanded form automatically also included the bmi30 and smoker variables as well as the interaction.
Based on a bit of subject matter knowledge of how medical costs may be related to patient characteristics, we developed what we think is a more accurately-specified regression formula. To summarize the improvements, we:
- Added a non-linear term for age
- Created an indicator for obesity
- Specified an interaction between obesity and smoking
We'll train the model using the lm() function as before, but this time we'll add the newly constructed variables and the interaction term:
> ins_model2 <- lm(charges ~ age + age2 + children + bmi + sex + bmi30*smoker + region, data = insurance)
Next, we summarize the results:
> summary(ins_model2)

The model fit statistics help to determine whether our changes improved the performance of the regression model. Relative to our first model, the R-squared value has improved from 0.75 to about 0.87. Our model is now explaining 87 percent of the variation in medical treatment costs. Additionally, our theories about the model's functional form seem to be validated. The higher-order age2 term is statistically significant, as is the obesity indicator, bmi30. The interaction between obesity and smoking suggests a massive effect; in addition to the increased costs of over $13,404 for smoking alone, obese smokers spend another $19,810 per year. This may suggest that smoking exacerbates diseases associated with obesity.
