Chapter 20. The Basics of Machine Learning in OpenCV
In this chapter, we’ll begin a discussion of the machinery that is used to turn vision into perception—in other words, the machinery that turns the visual inputs into meaningful visual semantics.
In the previous chapters we have discussed how to turn 2D or 2D+3D sensor information into features, clusters, or geometric information. In the next three chapters, we’ll use the results of these techniques to turn features, segmentations, and their geometry into recognition of scenes or objects; it is this step that turns raw information into a percept: what the machine is seeing and where it is relative to the camera.
In this chapter we will cover the basics of machine learning, focusing mainly on what it is. We will look at some simple machine learning capabilities of the library that form a good starting point for understanding the basic ideas in machine learning as a whole. In the next chapter, we will get into more detail about how modern machine learning methods are implemented in the library.1
What Is Machine Learning?
The goal of machine learning (ML)2 is to turn data into information. After learning from a collection of data, we want a machine to be able to answer questions about the data: What other data is most similar to this data? Is there a car in the image? What ad will the user respond to? There is often a cost component, so this question could become: “Of our most profitable products, which one will the user most likely buy if we show them an ad for it?” Machine learning turns data into information by extracting rules or patterns from that data.
Training and Test Sets
The sort of machine learning we are interested in works on raw numerical data, such as temperature values, stock prices, and color intensities. The data is often preprocessed into features.3 We might, for example, take a database of 10,000 face images, run an edge detector on the faces, and then collect features such as edge direction, edge strength, and offset from face center for each face. We might obtain 500 such values per face or a feature vector of 500 entries. We could then use machine learning techniques to construct some kind of model from this collected data. If we want to see only how faces fall into different groups (wide, narrow, etc.), then a clustering algorithm would be the appropriate choice. If we want to learn to predict the age of a person from, for example, the pattern of edges detected on his or her face, then a classifier algorithm would be appropriate. To meet our goals, machine learning algorithms analyze our collected features and adjust weights, thresholds, and other model parameters to maximize performance according to those goals. This process of parameter adjustment to meet a goal is what we mean by the term learning.
It is always important to know how well machine learning methods are working, and this can be a subtle task. Traditionally, one breaks up the available data set into a large training set (perhaps 9,000 faces, in our example) and a smaller test set (the remaining 1,000 faces). We can then run our classifier over the training set to learn our age prediction model given the data feature vectors. When we are done, we can test the age prediction classifier on the remaining images in the test set.
The test set is not used in training, and we do not let the classifier “see” the test set age labels. Only after training, we run the classifier over each of the 1,000 faces in the test set of data and record how well the ages it predicts (based on the feature vector) match the actual ages. If the classifier does poorly, we might try adding new features to our data or consider a different type of classifier. We’ll see in this chapter that there are many kinds of classifiers and many algorithms for training them.
If the classifier does well, we now have a potentially valuable model that we can deploy on data in the real world. Perhaps this system will be used to set the behavior of a video game based on age. As the person prepares to play, his or her face will be processed into 500 features (edge direction, edge strength, offset from face center, etc.). This data will be passed to the classifier; the age it returns will set the game play behavior accordingly. After it has been deployed, the classifier sees faces that it never saw before and makes decisions according to what it learned on the training set.
Finally, when developing a classification system, we often use a validation data set. Sometimes, testing the whole system at the end is too big a step to take. We often want to tweak parameters along the way before submitting our classifier to final testing. We might do this by breaking our example 10,000-face data set into three parts: a training set of 8,000 faces, a validation set of 1,000 faces, and a test set of 1,000 faces. Now, while we’re running through the training data set, we can “sneak” pretests on the validation data to see how we are doing. Only when we are satisfied with our performance on the validation set do we run the classifier on the test set for final judgment.
Note
It might strike you that you could do better than to train with 8,000 examples and validate on 1,000 in a 9,000-example training set. If so, you would be correct. The standard practice in such cases is actually to repeat this partitioning multiple times. In this case, nine times would make the most sense. In each case you would set aside a different group of 1,000 points to use for validation and train on the remaining 8,000. This process is called k-fold cross-validation, with k-fold meaning “done in k variations”—in our example, nine.
Supervised and Unsupervised Learning
Data sometimes has no labels; for example, we might just want to see what kinds of groups faces naturally form based on our edge-detection information. Sometimes the data has labels, such as the age of the person featured in each image. What this means is that machine learning data may be supervised (i.e., may utilize a teaching “signal” or “label” that goes with the data feature vectors), or it may be unsupervised, in which case the machine learning algorithm has no access to such labels and is expected to figure out the structure of the data on its own (see Figure 20-1).

Figure 20-1. Machine learning includes two broad subcategories: supervised and unsupervised learning. Supervised learning itself has two important subcomponents: classification and regression
Supervised learning can be categorical, such as learning to associate a name with a face, or the data can have numeric or ordered labels, such as age. When the data has names (categories) as labels, we say we are doing classification. When the data is numeric, we say we are doing regression: trying to fit a numeric output given some categorical or numeric input data.
Supervised learning also comes in shades of gray: it can involve one-to-one pairing of labels with data vectors or it may consist of reinforcement learning (sometimes called deferred learning). In reinforcement learning, the data label (also called the reward or punishment) can come long after the individual data vectors were observed. When a mouse is running down a maze to find food, the mouse may face a series of turns before it finally finds the food, its reward. That reward must somehow cast its influence back on all the sights and actions that the mouse took before finding the food. Reinforcement learning works the same way: the system receives a delayed signal (a reward or a punishment) and tries to infer a behavior (formally called a policy) for future runs. In this case, the policy being learned is actually the way of making decisions; e.g., which way to go at each step through the maze. Supervised learning can also have partial labeling, where some labels are missing (this is also called semisupervised learning), or it might have noisy labels, where some of the supplied labels are just wrong. Most ML algorithms handle only one or two of the situations just described. For example, the ML algorithms might handle classification but not regression; the algorithm might be able to do semisupervised learning but not reinforcement learning; the algorithm might be able to deal with numeric but not categorical data; and so on.
In contrast, often we don’t have labels for our data and are interested in seeing whether the data falls naturally into groups. The algorithms for this kind of unsupervised learning are often called clustering algorithms. In this situation, the goal is to group unlabeled data vectors that are “close” (in some predetermined or possibly even learned sense). We might just want to see how faces are distributed: do they form clumps of thin, wide, long, or short faces? If we’re looking at cancer data, do some cancers cluster into groups having different chemical signals? Unsupervised clustered data is also often used to form a feature vector for a higher-level supervised classifier. We might first cluster faces into face types (wide, narrow, long, short) and then use that as an input, perhaps with other data, such as average vocal frequency, to predict the gender of a person.
These two common machine learning tasks, classification and clustering, overlap with two of the most common tasks in computer vision, recognition and segmentation. This is sometimes referred to as the “what” and the “where.” That is, we often want our computer to name the object in an image (recognition, or “what”) and also to say where the object appears (segmentation, or “where”). Because computer vision makes such heavy use of machine learning, OpenCV includes many powerful machine learning algorithms in the ML libraries, located in the .../opencv/modules/ml and the .../opencv/modules/flann directories.
Note
The OpenCV machine learning code is general; that is, although it is highly useful for vision tasks, the code itself is not specific to vision. One could learn, say, genomic sequences using the appropriate routines. Of course, our concern here is mostly with object recognition given feature vectors derived from images.
Generative and Discriminative Models
Many algorithms have been devised to perform classification and clustering. OpenCV supports some of the most useful currently available statistical approaches to machine learning. Probabilistic approaches to machine learning, such as Bayesian networks or graphical models, are less well supported in OpenCV, partly because they are newer and still under active development. OpenCV tends to support discriminative algorithms, which give us the probability of the label given the data, P(L|D), rather than generative algorithms, which give the distribution of the data given the label, P(D|L). Although the distinction is not always clear, discriminative models are good for yielding predictions given the data, while generative models are good for giving you more powerful representations of the data or for conditionally synthesizing new data (think of “imagining” an elephant; you’d be generating data given a condition “elephant”).
It is often easier to interpret a generative model because it models (correctly or incorrectly) the cause of the data. Discriminative learning often comes down to making a decision based on some threshold that may seem arbitrary. For example, suppose a patch of road is identified in a scene partly because its color “red” is less than 125. But does this mean that red = 126 is definitely not road? Such issues can be hard to interpret. With generative models you are usually dealing with conditional distributions of data given the categories, so you can develop a feel for what it means to be “close” to the resulting distribution.
OpenCV ML Algorithms
The machine learning algorithms included in OpenCV are given in Table 20-1. Many of the algorithms listed are in the ML module; Mahalanobis and K-means are in the core module; face detection and object detection methods are in the objdetect module; FLANN takes a dedicated module, flann; and other algorithms are placed in opencv_contrib.
| Algorithm | Comment |
|---|---|
| Mahalanobis | A distance measure that accounts for the “stretchiness” of the data space by dividing out the covariance of the data. If the covariance is the identity matrix (identical variance), then this measure is identical to the Euclidean distance measure [Mahalanobis36]. |
| K-means | An unsupervised clustering algorithm that represents a distribution of data using K centers, where K is chosen by the user. The difference between this algorithm and expectation maximization (described shortly) is that here the centers are not Gaussian and the resulting clusters look more like soap bubbles, since centers (in effect) compete to “own” the closest data points. These cluster regions are often used as sparse histogram bins to represent the data. Invented by Steinhaus [Steinhaus56], as used by Lloyd [Lloyd57]. |
| Normal/Naïve Bayes classifier | A generative classifier in which features are assumed to be Gaussian distributed and statistically independent from one another, a strong assumption that is generally not true. For this reason, it’s often called a “naïve Bayes” classifier. However, this method often works surprisingly well. Original mention [Maron61; Minsky61]. |
| Decision trees | A discriminative classifier. The tree finds one data feature and a threshold at the current node that best divides the data into separate classes. The data is split and we recursively repeat the procedure down the left and right branches of the tree. Though not usually the top performer, it’s often the first thing you should try because it is fast and has high functionality [Breiman84]. |
| Expectation maximization (EM) | A generative unsupervised algorithm that is used for clustering. It will fit N multidimensional Gaussians to the data, where N is chosen by the user. This can be an effective way to represent a more complex distribution with only a few parameters (means and variances). Often used in segmentation. Compare with K-means listed previously [Dempster77]. |
| Boosting | A discriminative group of classifiers. The overall classification decision is made from the combined weighted classification decisions of the group of classifiers. In training, we learn the group of classifiers one at a time. Each classifier in the group is a “weak” classifier (only just above chance performance). These weak classifiers are typically composed of single-variable decision trees called stumps. In training, the decision stump learns its classification decisions from the data and also learns a weight for its “vote” from its accuracy on the data. Between training each classifier one by one, the data points are reweighted so that more attention is paid to data points where errors were made. This process continues until the total error over the data set, arising from the combined weighted vote of the decision trees, falls below a set threshold. This algorithm is often effective when a large amount of training data is available [Freund97]. |
| Random trees | A discriminative forest of many decision trees, each built down to a large or maximal splitting depth. During learning, each node of each tree is allowed to choose splitting variables only from a random subset of the data features. This helps ensure that each tree becomes a statistically independent decision maker. In run mode, each tree gets an unweighted vote. This algorithm is often very effective and can also perform regression by averaging the output numbers from each tree implemented [Ho95; Criminisi13; Breiman01]. |
| K-nearest neighbors | The simplest possible discriminative classifier. Training data are simply stored with labels. Thereafter, a test data point is classified according to the majority vote of its K nearest other data points (in a Euclidean sense of nearness). This is probably the simplest thing you can do. It is often effective but it is slow and requires lots of memory [Fix51], but see the FLANN entry. |
| Fast Approximate Nearest Neighbors (FLANN)a | OpenCV includes a full fast approximate nearest neighbor library developed by Marius Muja [Muja09]. This allows fast approximations to nearest neighbor and K nearest neighbor matching. |
| Support vector machine (SVM) | A discriminative classifier that can also do regression. A distance function between any two data points in a higher-dimensional space is defined. (Projecting data into higher dimensions makes the data more likely to be linearly separable.) The algorithm learns separating hyperplanes that maximally separate the classes in the higher dimension. It tends to be among the best with limited data, losing out to boosting or random trees only when large data setsb are available [Vapnik95]. |
| Face detector/cascade classifier | An object detection application based on a clever use of boosting. The OpenCV distribution comes with a trained frontal face detector that works remarkably well. You may train the algorithm on other objects with the software provided. You may also use other features or create your own features for this classifier. It works well for rigid objects and characteristic views. After its inventors, this classifier is also commonly known as a “Viola-Jones Classifier” [Viola04]. |
| Waldboost | A derivative of the cascade method of Viola (see the preceding entry), Waldboost is an object detector that is very fast and often outperforms the traditional cascade classifier for a variety of tasks [Sochman05]. It is in .../opencv_contrib/modules. |
| Latent SVM | The Latent SVM method uses a parts-based model to identify composite objects based on recognizing the individual components of the object and learning a model of how those components should expect to be found relative to one another [Felzenszwalb10]. |
| Bag of Words | The Bag of Words method generalizes techniques heavily used in document classification to visual image classification. This method is powerful because it can be used to identify not only individual objects, but often scenes and environments as well. |
a Since you are wondering, the L in FLANN stands for library (FLANN = Fast Library for Approximate Nearest Neighbors). b What is “large data”? There is no answer, since it depends on how fast the underlying generating process changes. But here’s a very crude rule of thumb: 10 data points per category/object per meaningful dimension (feature). So, two classes, three dimensions needs at least 2*10*10*10 = 2,000 data points to be “large” for that problem. | |
Using Machine Learning in Vision
In general, all the algorithms in Table 20-1 take as input a data vector made up of many features, where the number of features might well be in the thousands. Suppose your task is to recognize a certain type of object—for example, a person. The first problem that you will encounter is how to collect and label training data that falls into positive (there is a person in the scene) and negative (no person) cases. You will soon realize that people appear at different scales: their image may consist of just a few pixels, or you may be looking at an ear that fills the whole screen. Even worse, people will often be occluded: a man inside a car; a woman’s face; one leg peeking from behind a tree. You need to define what you actually mean by saying a person is in the scene.
Next, you have the problem of collecting data. Do you collect it from a security camera, go to photo-sharing websites and attempt to find “person” labels, or both (and more)? Do you collect movement information? Do you collect other information, such as whether a gate in the scene is open, the time, the season, the temperature? An algorithm that finds people on a beach might fail on a ski slope. You need to capture the variations in the data: different views of people, lightings, weather conditions, shadows, and so on.
After you have collected lots of data, how will you label it? You must first decide what you mean by “label.” Do you want to know where the person is in the scene? Are actions (running, walking, crawling, following) important? You might end up with a million images or more. How will you label all that? There are many tricks, such as doing background subtraction in a controlled setting and collecting the segmented foreground humans who come into the scene. You can use data services to help in classification; for example, you can pay people to label your images through Amazon’s Mechanical Turk. If you arrange tasks to be simple, you can get the cost down to somewhere around a penny per label. Finally, you may use GPUs and/or clusters of computers to render objects/people/faces/hands using computer graphics. Using camera model parameters, you can often generate very realistic images where ground truth is known since you generated the data.
After labeling the data, you must decide which features to extract from the objects. Again, you must know what you are after. If people always appear right side up, there’s no reason to use rotation-invariant features and no reason to try to rotate the objects beforehand. In general, you must find features that express some invariance in the objects, such as scale-tolerant histograms of gradients or colors or the popular SIFT features.4 If you have background scene information, you might want to first remove it to make other objects stand out. You then perform your image processing, which may consist of normalizing the image (rescaling, rotation, histogram equalization, etc.) and computing many different feature types. The resulting data vectors are each given the label associated with that object, action, or scene.
Once the data is collected and turned into feature vectors, you often want to break up the data into training, (possibly) validation, and test sets. As we saw earlier, is a “best practice” to do your learning, validation, and testing within a cross-validation framework. Recall that there the data is divided into K subsets and you run many training (possibly validation) and test sessions, where each session consists of different sets of data taking on the roles of training (validation) and test.5 The test results from these separate sessions are then averaged to get the final performance result. Cross-validation gives a more accurate picture of how the classifier will perform when deployed in operation on novel data. (We’ll have more to say about this in what follows.)
Now that the data is prepared, you must choose your classifier. Often this choice is dictated by computational, data, or memory considerations. For some applications, such as online user preference modeling, you must train the classifier rapidly. In this case, nearest neighbors, normal Bayes, or decision trees would be a good choice. If memory is a consideration, decision trees or neural networks are space efficient. If you have time to train your classifier but it must run quickly, neural networks are a good choice, as are naïve Bayes classifiers and support vector machines. If you have time to train and some time to run, but need high accuracy, then boosting and random trees are likely to fit your needs. If you just want an easy, understandable sanity check that your features are chosen well, then decision trees or nearest neighbors are good bets. For best “out of the box” classification performance, try boosting or random trees first.
Note
There is no “best” classifier (see http://en.wikipedia.org/wiki/No_free_lunch_theorem). Averaged over all possible types of data distributions, all classifiers perform the same. Thus, we cannot say which algorithm in Table 20-1 is the “best.” Over any given data distribution or set of data distributions, however, there is a best classifier. Thus, when faced with real data it’s a good idea to try many classifiers. Consider your purpose: Is it just to get the right score, or is it to interpret the data? Do you seek fast computation, small memory requirements, or confidence bounds on the decisions? Different classifiers have different properties along these dimensions.
Variable Importance
Two of the algorithms in Table 20-1 allow you to assess a variable’s importance.6 Given a vector of features, how do you determine the importance of those features for classification accuracy? Binary decision trees do this directly: you train them by selecting which variable best splits the data at each node. The top node’s variable is the most important variable; the next-level variables are the second most important, and so on.7 Random trees can measure variable importance using a technique developed by Leo Breiman [Breiman02]; this technique can be used with any classifier, but so far it is implemented only for decision and random trees in OpenCV.
One use of variable importance is to reduce the number of features your classifier must consider. Starting with many features, you train the classifier and then find the importance of each feature relative to the other features. You can then discard unimportant features. Eliminating unimportant features improves speed (since it eliminates the processing it took to compute those features) and makes training and testing quicker. Also, if you don’t have enough data, which is often the case, then eliminating unimportant variables can increase classification accuracy; this yields faster processing with better results.
Breiman’s variable importance algorithm runs as follows.
-
Train a classifier on the training set.
-
Use a validation or test set to determine the accuracy of the classifier.
-
For every data point and a chosen feature, randomly choose a new value for that feature from among the values the feature has in the rest of the data set (called “sampling with replacement”). This ensures that the distribution of that feature will remain the same as in the original data set, but now the actual structure or meaning of that feature is erased (because its value is chosen at random from the rest of the data).8
-
Train the classifier on the altered set of training data and then measure the accuracy of classification on the altered test or validation data set. If randomizing a feature hurts accuracy a lot, then that feature is very important. If randomizing a feature does not hurt accuracy much, then that feature is of little importance and is a candidate for removal.
-
Restore the original test or validation data set and try the next feature until we are done. The result is an ordering of each feature by its importance.
This procedure is built into random trees and decision trees. Thus, you can use random trees or decision trees to decide which variables you will actually use as features; then you can use the slimmed-down feature vectors to train the same (or another) classifier.
Diagnosing Machine Learning Problems
Getting machine learning to work well can be more of an art than a science. Algorithms often “sort of” work but not quite as well as you need them to. That’s where the art comes in; you must figure out what’s going wrong in order to fix it. Although we can’t go into all the details here, we’ll give an overview of some of the more common problems you might encounter.9
First, some rules of thumb: more data beats less data, and better features beat better algorithms. If you design your features well—maximizing their independence from one another and minimizing how they vary under different conditions—then almost any algorithm will work well. Beyond that, there are three common problems:
- Bias
- Your model assumptions are too strong for the data, so the model won’t fit well.
- Variance
- Your algorithm has memorized the data including the noise, so it can’t generalize.
- Bugs
- It is not uncommon for machine learning code that contains seemingly severe bugs to “learn its way around” the bugs, often yielding only degraded performance when absolute failure would be expected.
Figure 20-2 shows the basic setup for statistical machine learning. Our job is to model the true function f that transforms the underlying inputs to some output. This function may be a regression problem (e.g., predicting a person’s age from their face10) or a category prediction problem (e.g., identifying a person given their facial features). For problems in the real world, noise and unconsidered effects can cause the observed outputs to differ from the theoretical outputs. For example, in face recognition we might learn a model of the measured distance between eyes, mouth, and nose to identify a face. But lighting variations from a nearby flickering bulb might cause noise in the measurements, or a poorly manufactured camera lens might cause a systematic distortion in the measurements that wasn’t considered as part of the model. These effects will cause accuracy to suffer.

Figure 20-2. Setup for statistical machine learning: we train a classifier to fit a data set; the true model f is almost always corrupted by noise or unknown influences
Figure 20-3 shows model bias (resulting in underfitting) and variance (resulting in overfitting) of data in the upper two panels and the consequences in terms of error with training set size in the lower two panels. On the left side of Figure 20-3 we attempt to train a classifier to predict the data in the lower panel of Figure 20-2. If we use a model that’s too restrictive—indicated here by the heavy, straight dashed line—then we can never fit the underlying true parabola f indicated by the thinner dashed line. Thus, the fit to both the training data and the test data will be poor, even with a lot of data. In this case we have bias because both training and test data are predicted poorly. On the right side of Figure 20-3 we fit the training data exactly, but this produces a nonsense function that fits every bit of noise. Thus, it memorizes the training data as well as the noise in that data. Once again, the resulting fit to the test data is poor. Low training error combined with high test error indicates a variance (overfit) problem.

Figure 20-3. Poor model fitting in machine learning and its effect on training and test prediction performance, where the true function is graphed by the lighter dashed line at top: a biased (underfit) model for the data (upper left) yields high error in predicting the training and the test set (lower left), whereas a variance (overfit) model for the data (upper right) yields low error in the training data but high error in the test data (lower right)
Sometimes you have to be careful that you are solving the correct problem. If your training and test set error are low but the algorithm does not perform well in the real world, the data set may have been chosen from unrealistic conditions—perhaps because these conditions made collecting or simulating the data easier. If the algorithm just cannot reproduce the test or training set data, then perhaps the algorithm is the wrong one to use, the features that were extracted from the data are ineffective, or the “signal” just isn’t in the data you collected. Table 20-2 lays out some possible fixes to the problems we’ve described here. Of course, this is not a complete list of the possible problems or solutions. It takes careful thought and design of what data to collect and what features to compute in order for machine learning to work well. It can also take some systematic thinking to diagnose machine learning problems.
| Problem | Possible solutions |
|---|---|
| Bias |
|
| Variance |
|
| Good test/train, bad real world |
|
| Model can’t learn test or train |
|
Cross-validation, bootstrapping, ROC curves, and confusion matrices
Finally, there are some basic tools that are used in machine learning to measure results. In supervised learning, one of the most basic problems is simply knowing how well your algorithm has performed: how accurate is it at classifying or fitting the data? You might think: “Easy, I’ll just run it on my test or validation data and get the result.” But for real problems, we must account for noise, sampling fluctuations, and sampling errors. Simply put, your test or validation set of data might not accurately reflect the actual distribution of data. To get closer to “guessing” the true performance of the classifier, we employ the technique of cross-validation and/or the closely related technique of bootstrapping.11
Recall that, in its most basic form, cross-validation involves dividing the data into K different subsets of data. You train on K – 1 of the subsets and test on the final subset of data (the “validation set”) that wasn’t trained on. You do this K times, where each of the K subsets gets a “turn” at being the validation set, and then average the results.
Bootstrapping is similar to cross-validation, but the validation set is selected at random from the training data. Selected points for that round are used only in test, not training. Then the process starts again from scratch. You do this N times, where each time you randomly select a new set of validation data and average the results in the end. Note that this means some and/or many of the data points are reused in different validation sets, but the results are often superior compared to cross-validation.12
Using either one of these techniques can yield more accurate measures of actual performance. This increased accuracy can in turn be used to tune parameters of the learning system as you repeatedly change, train, and measure.
Two other immensely useful ways of assessing, characterizing, and tuning classifiers are plotting the receiver operating characteristic (ROC, often pronounced “rock”) curve and the confusion matrix; see Figure 20-4. The ROC curve measures the response of the performance parameter of the classifier over the full range of settings of that parameter (each point on a ROC curve may also be computed with cross-validation). Let’s say the parameter is a threshold. Just to make this more concrete, suppose we are trying to recognize yellow flowers in an image and that we have a threshold on the color yellow as our detector. Setting the yellow threshold extremely high would mean that the classifier would fail to recognize any yellow flowers, yielding a false positive rate of 0 but at the cost of a true positive rate also at 0 (lower-left part of the curve in Figure 20-4). On the other hand, if the yellow threshold is set to 0, then any signal at all counts as a recognition. This means that all of the true positives (the yellow flowers) are recognized as well as all the false positives (orange and red flowers); thus, we have a false positive rate of 100% (upper-right part of the curve in Figure 20-4). The best possible ROC curve would be one that follows the y-axis up to 100% and then cuts horizontally over to the upper-right corner. Failing that, the closer the curve comes to the upper-left corner, the better. In practice, one often computes the fraction of area under the ROC curve versus the total area of the ROC plot as a summary statistic of merit: the closer that ratio is to 1, the better is the classifier.13

Figure 20-4. ROC curve and associated confusion matrix: the former shows the response of correct classifications to false positives along the full range of varying a performance parameter of the classifier; the latter shows the false positives (false recognitions) and false negatives (missed recognitions)
Figure 20-4 also shows a confusion matrix—a chart of true and false positives along with true and false negatives. It is another quick way to assess the performance of a classifier: ideally we’d see 100% along the diagonal and 0% elsewhere. If we have a classifier that can learn more than one class (e.g., a multilayer perceptron or random forest classifier, which can learn many different class labels at once), then the confusion matrix is generalized to cover all class labels, and the entries represent the overall percentages of true and false positives and negatives over all class labels. The ROC curve for multiple classes just records the true and false decisions over the test data set.
Cost of misclassification
One thing we haven’t discussed much here is the cost of misclassification. That is, if our classifier is built to detect poisonous mushrooms (we’ll see an example that uses just such a data set in the next chapter), then we are willing to have more false negatives (edible mushrooms mistaken as poisonous) as long as we minimize false positives (poisonous mushrooms mistaken as edible). The ROC curve can help with this; we can set our ROC parameter to choose an operation point lower on the curve—toward the lower left of the graph in Figure 20-4. The other way of doing this is to weight false positive errors more than false negatives when generating the ROC curve. For example, you can set the cost of each false positive error to be equal to 10 false negatives.14 With some OpenCV machine learning algorithms, such as decision trees and SVM, you can regulate this balance of “hit rate versus false alarm” by specifying prior probabilities of the classes themselves (which classes are expected to be more likely and which less) or by specifying weights of the individual training samples.
Mismatched feature variance
Another common problem with training some classifiers arises when the feature vector comprises features of widely different variances. For instance, if one feature is represented by lowercase ASCII characters, then it ranges over only 26 different values. In contrast, a feature that is represented by the count of biological cells on a microscope slide might vary over several billion values. An algorithm such as K-nearest neighbors might then see the first feature as relatively constant (nothing to learn from) compared to the cell-count feature. The way to correct this problem is to preprocess each feature variable by normalizing for its variance. This practice is acceptable provided the features are not correlated with each other; when features are correlated, you can normalize by their average variance or by their covariance. Some algorithms, such as decision trees,15 are not adversely affected by widely differing variance and so this precaution need not be taken. A rule of thumb is that if the algorithm depends in some way on a distance measure (e.g., weighted values), then you should normalize for variance. One may normalize all features at once and account for their covariance by using the Mahalanobis distance, which is discussed later in this chapter. Readers familiar with machine learning or signal processing might recognize this as the technique called “whitening” the data.
We now turn to discussing some of the machine learning algorithms supported in OpenCV.
Legacy Routines in the ML Library
Much of the ML library uses a common C++ interface that is object based and implements each algorithm inside of an object derived from this common base class. In this way, the access to the algorithms and their usage is standardized across the library. However, some of the most basic methods in the library do not conform to this standard interface because they were implemented in the early days of the library when the common interface had not yet been designed.16
Because it is the most basic methods that do not conform to the object interface, we will look at these first. In the next chapter, we will move on to the object interface and the many algorithms implemented through it. For now, the methods we will cover are K-means clustering, the Mahalanobis distance (and its utility in the context of K-means clustering), and finally a partitioning technique that is normally used as a means of improving both the speed and the accuracy of K-means clustering.
K-Means
K-means attempts to find the natural clusters in a set of vector-valued data. The user sets the desired number of clusters and then the K-means algorithm rapidly finds a good placement for the centers of those clusters. Here, “good” means that the cluster centers tend to end up located in the middle of the natural clumps of data. The K-means algorithm is one of the most used clustering techniques and has strong similarities to the Expectation Maximization algorithm17 as well as some similarities to the mean-shift algorithm discussed in Chapter 17 (implemented as cv::meanShift() in the CV library). K-means is an iterative algorithm and, as implemented in OpenCV, is also known as Lloyd’s algorithm [Lloyd82] or (equivalently) “Voronoi iteration.” The algorithm runs as follows.
-
Take as input a data set D and desired number of clusters K (chosen by the user).
-
Randomly assign (sufficiently separated) cluster center locations.
-
Associate each data point with its nearest cluster center.
-
Move cluster centers to the centroid of their data points.
-
Return to Step 3 until convergence (i.e., centroid does not move).
Figure 20-5 shows K-means in action; in this case, it takes just three iterations to converge. In real cases the algorithm often converges rapidly, but there are still times when it will require a large number of iterations.

Figure 20-5. K-means in action for three iterations: (a) cluster centers are placed randomly and each data point is then assigned to its nearest cluster center; (b) cluster centers are moved to the centroid of their points; (c) data points are again assigned to their nearest cluster centers; (d) cluster centers are again moved to the centroid of their points
Problems and solutions
K-means is an extremely effective clustering algorithm, but it does have three important shortcomings:
-
It isn’t guaranteed to find the best possible solution to locating the cluster centers. However, it is guaranteed to converge to some solution (i.e., the iterations won’t continue indefinitely).
-
It doesn’t tell you how many cluster centers you should use. If we had chosen two or four clusters for the example in Figure 20-5, then the results would be different and perhaps less than intuitive.
-
It presumes that the covariance in the space either doesn’t matter or has already been normalized (we will return to this, and the Mahalanobis distance, shortly).
Each one of these problems has a “solution,” or at least an approach that helps. The first two of these solutions depends on “explaining the variance of the data.” In K-means, each cluster center “owns” its data points and we compute the variance of those points.18
The best clustering minimizes the variance without causing too much complexity (too many clusters). With that in mind, we can ameliorate the listed problems as follows:
-
Run K-means several times, each with different placement of the cluster centers; then choose the run whose results exhibit the least variance. OpenCV can do this automatically; you need only specify the number of such clustering attempts (see the
attemptsparameter ofcv::kmeans). -
Start with one cluster and try an increasing number of clusters (up to some limit), each time employing the method of Step 1 as well. Usually the total variance will shrink quite rapidly, after which an “elbow” will appear in the variance curve; this indicates that a new cluster center does not significantly reduce the total variance. Stop at the elbow and keep that many cluster centers.
-
Multiply the data by the inverse covariance matrix (as described in the section “Mahalanobis Distance”). For example, if the input data vectors D are organized as rows with one data point per row, then normalize the “stretch” in the space by computing a new data vector
 , where
, where  .
.
K-means code
The call for K-means is simple:
double cv::kmeans( // returns (best) compactness cv::InputArray data, // Your data, in a float type int K, // Number of clusters cv::InputOutputArray bestLabels, // Result cluster indices (int's) cv::TermCriteria criteria, // iterations and/or min dist int attempts, // starts to search for best fit int flags, // initialization options cv::OutputArray centers = cv::noArray() // (optional) found centers );
The data array is a matrix of multidimensional data points, one per row, where each element of the data is a regular floating-point value (i.e., CV_32FC1). Alternatively, data may be simply a single column of entries, each of which is a multidimensional point (i.e., of type CV_32FC2 or CV_32FC3 or even CV_32FC(M)).19 The parameter K is the number of clusters you want to find, and the return vector bestLabels contains the final cluster index for each data point. In the case of criteria, you may specify either the maximum number of iterations you would like the algorithm to run, or the small distance that will be used to determine when a cluster center is effectively stationary (i.e. if it moves less than the given small distance). Of course, you can specify both of these criteria as well.
The parameter attempts tells cv::kmeans() to automatically run some number of times, each time starting with a new set of seed points, and to keep only the best result. The quality of the results is gauged by the compactness—that is, the sum of squared distances between every point and the center of the cluster to which that point was associated.
The flags parameter may be any of the following values (with the first being the default): cv::KMEANS_RANDOM_CENTERS, cv::KMEANS_USE_INITIAL_LABELS, or cv::KMEANS_PP_CENTERS. In the case of cv::KMEANS_RANDOM_CENTERS, we assign the starting cluster centers as described earlier, by randomly selecting from the points in the data set. In the case of cv::KMEANS_USE_INITIAL_LABELS, the values stored in the parameter bestLabels at the time the function is called will be used to compute the initial cluster centers. Finally, the cv::KMEANS_PP_CENTERS option instructs cv::kmeans() to use the method of Sergei Vassilvitskii and David Arthur [Arthur07] called K-means++ to assign the cluster centers. The details of this method are not critical to us here, but what is important is that this method more prudently chooses the starting points for the cluster centers and typically gives better results in fewer iterations than the default method. In modern applications, K-means++ is increasingly the standard being used.
Note
Even though, in theory, the K-means algorithm can behave rather badly, it is heavily used in practice because most of the time it does quite well. The fact that, in the worst case, the cluster assignment problem is NP-hard means that you are unlikely to ever get a truly optimal answer, but for most applications, a “good” answer is good enough. Still, there are some unsettling problems with the K-means algorithm. One of them is that, in certain cases, it can be tricked into producing what mathematicians call “arbitrarily bad” results. This means that no matter how wrong you might fear the answer might be, some tricky person can come up with a circumstance in which it will be at least that wrong, or worse. As a result there has been some interest in past years (and continues to be) in techniques that provide either some general improvement in performance, or in the best case, some provable bounds that can give the user a little more confidence in the results. One such algorithm is K-means++.
Finally, on completion, the computed centers for the clusters will be placed into the array centers. You can omit centers if you do not require them (in this case by passing cv::noArray()). The function always returns the computed compactness.
It’s instructive to see a complete example of K-means in code (Example 20-1). An added benefit of the example is that the data-generation sections can be used to test other machine learning routines as well.
Example 20-1. Using K-means
#include "opencv2/highgui/highgui.hpp"
#include "opencv2/core/core.hpp"
#include <iostream>
using namespace cv;
using namespace std;
static void help( char* argv[] ) {
cout << "
This program demonstrates kmeans clustering.
"
" It generates an image with random points, then assigns a random number
"
" of cluster centers and uses kmeans to move those cluster centers to their
"
" representative location
"
"Usage:
"
<<argv[0] <<"
" << endl;
}
int main( int /*argc*/, char** /*argv*/ ) {
const int MAX_CLUSTERS = 5;
cv::Scalar colorTab[] = {
cv::Scalar( 0, 0, 255 ),
cv::Scalar( 0, 255, 0 ),
cv::Scalar( 255, 100, 100 ),
cv::Scalar( 255, 0, 255 ),
cv::Scalar( 0, 255, 255 )
};
cv::Mat img( 500, 500, CV_8UC3 );
cv::RNG rng( 12345 );
for(;;) {
int k, clusterCount = rng.uniform(2, MAX_CLUSTERS+1);
int i, sampleCount = rng.uniform(1, 1001);
cv::Mat points(sampleCount, 1, CV_32FC2), labels;
clusterCount = MIN(clusterCount, sampleCount);
cv::Mat centers(clusterCount, 1, points.type());
/* generate random sample from multigaussian distribution */
for( k = 0; k < clusterCount; k++ ) {
cv::Point center;
center.x = rng.uniform(0, img.cols);
center.y = rng.uniform(0, img.rows);
cv::Mat pointChunk = points.rowRange(
k*sampleCount/clusterCount,
k == clusterCount - 1 ? sampleCount : (k+1)*sampleCount/clusterCount
);
rng.fill(
pointChunk,
RNG::NORMAL,
cv::Scalar(center.x, center.y),
cv::Scalar(img.cols*0.05, img.rows*0.05)
);
}
randShuffle(points, 1, &rng);
kmeans(
points,
clusterCount,
labels,
cv::TermCriteria(
cv::TermCriteria::EPS | cv::TermCriteria::COUNT,
10,
1.0
),
3,
KMEANS_PP_CENTERS,
centers
);
img = Scalar::all(0);
for( i = 0; i < sampleCount; i++ ) {
int clusterIdx = labels.at<int>(i);
cv::Point ipt = points.at<cv::Point2f>(i);
cv::circle( img, ipt, 2, colorTab[clusterIdx], cv::FILLED, cv::LINE_AA );
}
cv::imshow("clusters", img);
char key = (char)waitKey();
if( key == 27 || key == 'q' || key == 'Q' ) // 'ESC'
break;
}
return 0;
}
In this code we used highgui to create a window output interface and include core.hpp because it contains cv::kmeans().20 The basic operation of the program is to first choose a number of clusters to generate, generate centers for those clusters, and then generate a cloud of points around the generated center. We will then come back and see whether cv::kmeans() can effectively rediscover this structure we put into the sample data. In main(), we first do some minor housekeeping, like setting up the coloring we will later use for displayed clusters. We then set up a main loop, which will let users run over and over again, generating different sets of test data.
This loop begins by determining how many clusters there will be in the underlying data and how many data points will be generated. Then for each cluster the center is generated and points are generated from a Gaussian distribution around that center. The points are then shuffled so that they will not be in cluster order.
At this point we turn the K-means algorithm loose on the data. In this example we don’t search the possible cluster counts, we just tell cv::kmeans() how many clusters there will be. The resulting labeling is computed and placed in labels.
The final for{} loop just draws the results. This is followed by deallocating the allocated arrays and displaying the results in the “clusters” image. Finally, we wait indefinitely (cv::waitKey(0)) to allow the user to do another run or to quit via the Esc key.
Mahalanobis Distance
We encountered the Mahalanobis distance earlier in Chapter 5 as a means of computing the distance between a point and a distribution center that was sensitive to the shape of the distribution. In the context of the K-means algorithm, the concept of the Mahalanobis distance can serve us in two different ways. The first application comes from understanding the Mahalanobis distance from an alternative point of view in which we regard it as measuring Euclidean distance on a deformed space. This allows us to use what we know about the Mahalanobis distance to create a rescaling of data that can substantially improve the performance of the K-means algorithm. The second application of the Mahalanobis distance is as a means of assigning novel data points to the clusters defined by the K-means algorithm.
Using the Mahalanobis distance to condition input data
In Example 20-1, we mentioned briefly the possibility that the data might be arranged in the space in a highly asymmetrical way. Of course, the entire point of using K-means is to assert that the data is clustered in a nonuniform way and to try to discover something about that clustering. However, there is an important distinction between “asymmetrical” and “nonuniform.” If, for example, all of your data is spread out a great distance in some dimensions and relatively little distance in others, then the K-means algorithm will behave poorly. An example of such a situation is shown in Figure 20-6.

Figure 20-6. The Mahalanobis computation allows us to reinterpret the data’s covariance as a “stretch” of the space: (a) the vertical distance between raw data sets is less than the horizontal distance; (b) after the space is normalized for variance, the horizontal distance between data sets is less than the vertical distance
Such situations often arise simply because there are different underlying units to different dimensions of a data vector. For example, if persons in a community are represented by their height, age, and the total number of years of schooling they have had, the simple fact that the units of height and age are different will result in a very different dispersion of the data those dimensions. Similarly, age and years of schooling, though they have the same units, have a naturally very different variance among natural populations.
This example, however, suggests a simple technique that can be very helpful. This technique is to look at the data set as a whole, and to compute the covariance matrix for the entire data set. Once this is done, we can rescale the entire data set using that covariance. By using such a technique, we can rescale the data in Figure 20-6(a) into something more like in Figure 20-6(b).
Recall that we encountered the Mahalanobis distance in Chapter 5 when we looked at the function cv::Mahalonobis(). The traditional use of the Mahalanobis distance is to measure the distance of a point from a distribution in such a way that the distance is measured in units of the distribution’s variance in the particular direction of the point. (For those familiar with the concept of the Z-score from statistics, the Mahalanobis distance is the generalization of the Z-score to multidimensional spaces.) We compute the Mahalanobis distance using the inverse of the covariance of the distribution:

where E[·] is the “expectation operator.” The actual formula for the Mahalanobis distance is then:

At this point, we can look at the situation in one of two ways: we can either say that we would like to use the Mahalanobis distance rather than the Euclidean distance in the K-means algorithm, or we can think of rescaling the data first and then using the Euclidean distance in the rescaled space. The first is probably a more intuitive way to look at things, but the second is much easier computationally—if only because we don’t want to actually crack open and modify the nice K-means implementation that is already provided for us. In the end, because the transformation is a linear one, either interpretation is possible.
After a little thought, it should be clear that we can simply rescale the data with the following operation:

Here D* is the set of new data vectors we will use, and D is the original data. The factor of Σ–1/2 is just the square root of the inverse covariance.21
In this case, we do not actually make direct use of cv::Mahalanobis(). Instead, we compute the set covariance using the cv::calcCovarMatrix(), invert it with cv::invert() (using cv::DECOMP_EIG22) and finally compute the square root.23
Using the Mahalanobis distance for classification
Given a set of cluster labels, from K-means clustering or any other method, we can use those labels to attempt to guess what cluster some novel point most likely belongs to. In the case where the clusters themselves are, or are thought to be, Gaussian distributed, it makes sense to apply the concept of the Mahalanobis distance also to this assignment problem.
The first step we need to take in order to make such assignments is to characterize each cluster in terms of its mean and covariance. Once we have done this, we can compute a Mahalanobis distance to each cluster center for any novel point.
From here you might guess that the point with the smallest Mahalanobis distance would be the winner, but it is not quite that simple. This would be true if all of the clusters had the same number of elements (or as a statistician would say, “if the prior probability of being a member of each cluster is equal”).24
This distinction is succinctly captured by Bayes’ rule, which states (in words) that, for two propositions A and B, the probability that A is true given B is (in general) not equal to the probability that B is true given A. In equation form, this looks like:
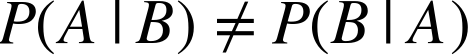
Instead, it says (again in words) that the probability that A is true given B multiplied by the probability that B is true in the first place is equal to the probability that B is true given A multiplied by the probability that A is true in the first place. In equation form, this looks like:

If you are trying to figure out how to tie this back to our Mahalanobis distance problem, remember that the Mahalanobis distance is telling us something about the probability that a particular sample came from a particular cluster, but—and here is the key point—that is the probability provided it came from that cluster at all. Seen a different way, we want to know the probability that our point is in cluster C given its value of ![]() . But the Mahalanobis distance is telling us the opposite, namely the probability of getting
. But the Mahalanobis distance is telling us the opposite, namely the probability of getting ![]() if we are in cluster C. Here it is written out as an equation (Bayes’s rule, slightly rearranged):
if we are in cluster C. Here it is written out as an equation (Bayes’s rule, slightly rearranged):

This means that in order to compare two Mahalanobis distances between two different clusters, we should take into account the sizes of the clusters. Given that the probability is asserted to be Gaussian for each cluster, the figure of merit that we should be comparing, called the likelihood, is:25

In this equation the ratio Nc divided by ND is the fraction of data points in cluster C relative to the total number of data points. This ratio is the prior probability of cluster C. The second term contains the inverse square root of the determinant of the covariance of cluster C, and the exponential factor that contains the squared Mahalanobis distance.
Summary
In this chapter we started with a basic discussion of what machine learning is and looked at which parts of that large problem space are addressed by the OpenCV library and which are not. We looked at the distinction between training and test data. We learned that generative models are those that attempt to find structure in existing data without supervision or labeled training data, while discriminative models are those that learn from examples and attempt to generalize from what they have been shown. We then moved on to look at two very fundamental tools that are available in the OpenCV library, K-means clustering and Mahalanobis distance. We saw how these could be used to build simple models and solve interesting problems. We note, in passing, that OpenCV now also supports deep neural networks, but that in their current form they are in the experimental opencv_contrib. That code is described in Appendix B (cnn_3dobj and dnn).
Exercises
-
If features in each data point vary widely in scale (say the first feature varies from 1 to 100 and the second feature varies from 0.0001 to 0.0002), explain whether and why it would pose a problem for:
-
SVM
-
Decision trees
-
Back-propagation
-
-
One way of removing the scale differences across features is to normalize the data. Two ways of doing this are to divide by the standard deviation of each feature or to divide by the maximum minus the minimum value. For each method of normalization, describe a set of data this would work well for and one that it would not work well for.
-
Consider the case of rescaling the input data set using the Mahalanobis distance before using the K-means algorithm. Prove that prescaling the data according to the formula
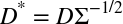 is exactly equivalent to modifying the algorithm to use the Mahalanobis distance internally.
is exactly equivalent to modifying the algorithm to use the Mahalanobis distance internally. -
Consider trying to learn the next stock price from several past stock prices. Suppose you have 20 years of daily stock data. Discuss the effects of various ways of turning your data into training and testing data sets. What are the advantages and disadvantages of the following approaches?
-
Take the even-numbered points as your training set and the odd-numbered points as your test set.
-
Randomly select points into training and test sets.
-
Divide the data in two, where the first half is for training and the second half for testing.
-
-
Divide the data into many small windows of several past points and one prediction point. Refer to Figure 20-3. Can you imagine conditions under which the test set error would be lower than the training set error?
-
Figure 20-3 was drawn for a regression problem. Label the first point on the graph A, the second point B, the third point A, the fourth point B, and so on. Draw a separation line for these two classes (A and B) that shows:
-
bias
-
variance
-
-
Refer to Figure 20-4.
-
Draw the generic best-possible ROC curve.
-
Draw the generic worst-possible ROC curve.
-
-
Draw a curve for a classifier that performs randomly on its test data.
-
Consider variable importance.
1 Note that machine learning, as with so many things, has been extended in the experimental opencv_contrib code as described in Appendix B. For more details, see the deep neural network repositories cnn_3dobj and dnn.
2 Machine learning is a vast topic. OpenCV deals mostly with statistical machine learning rather than subjects such as Bayesian networks, Markov random fields, and graphical models. Some good texts in machine learning include those by Hastie, Tibshirani, and Friedman [Hastie01]; Duda and Hart [Duda73]; Duda, Hart, and Stork [Duda00]; and Bishop [Bishop07]. For discussions on how to parallelize machine learning, see Ranger et al. [Ranger07] and Chu et al. [Chu07].
3 As of this writing, OpenCV does not include deep learning since, although promising, such techniques are still too new to know what to include. However, Convolutional Neural Networks [Fukushima80; LeCun98a; Ciresan11] are definitely a future candidate.
4 See Lowe’s SIFT keypoint feature demo.
5 One typically does the train (possibly validation) and test cycle 5 to 10 times.
6 This is commonly known as variable importance—meaning the importance of a variable, not importance that varies or fluctuates.
7 This technique by itself would be very sensitive to noise. What binary trees really do is build surrogate splits (other features to split on that result in almost the same decisions) for each node and compute the importance over all splits in all the nodes.
8 The actual implementation for random trees shuffles the feature values instead of generating completely new values.
9 Professor Andrew Ng at Stanford University gives the details in a web lecture entitled “Advice for Applying Machine Learning”.
10 An astute observer might note that since ages are likely to be reported as integers, this could be a classification problem rather than a regression problem. However, it is not the continuousness of the output that makes a problem a regression problem, it is the orderability of the output. Thus, even for integer ages, this is a regression problem.
11 For more information on these techniques, see “What Are Cross-Validation and Bootstrapping?”.
12 One of the main goals of bootstrapping is to find the optimal training parameters, because while it’s easy to average the training error, it may be nontrivial and/or inefficient to “average” several models into one.
13 It is worth noting that there are endless varieties of “figures of merit” for classifiers in the literature, and equally many reasons to prefer one over another. This particular one is relatively common, however, not because it is necessarily a better representation of the quality of a classifier, but because it is easily understood and well defined for almost any classification problem.
14 This is useful if you have some specific a priori notion of the relative cost of the two error types. For example, the cost of misclassifying one product as another in a supermarket checkout would be easy to quantify exactly beforehand.
15 Decision trees are not affected by variance differences in feature variables because each variable is searched only for effective separating thresholds. In other words, it doesn’t matter how large the variable’s range is as long as a clear separating value can be found.
16 At this time it is an open “to do” item of the library for someone to come along and write new interfaces to these legacy functions that conform to the object-based style used by the rest of MLL.
17 Specifically K-means is similar to the Expectation Maximization algorithm for Gaussian mixture models. This EM algorithm is also implemented in OpenCV (as cv::EM() in the ML library). We will encounter this algorithm later in the chapter.
18 In this context the variance of a point is the distance of the point from the cluster center. The variance of the points (plural) is typically the quadrature sum over the points. This sum is also called compactness.
19 Recall that this case is in fact exactly equivalent to an N × M matrix in which the N rows are the data points, the M columns are the individual components of each point’s location, and the underlying data type is cv::32FC1. Recall that, owing to the memory layout used for arrays, there is no distinction between these representations.
20 This is an example of the legacy issue again. cv::kmeans() predates the formal creation of the ML library, and so its prototype is in core.hpp rather than ml.hpp (as you probably imagined it would be).
21 If you are wondering why the inverse covariance is on the right in this formula, it is because the convention in the ML library is to represent a data set D as N rows of points and M columns for each point. Thus the data are rows and not columns.
22 DECOMP_SVD could also be used in this case, but it is somewhat slower and less accurate than DECOMP_EIG. DECOMP_EIG, even if it is slower than DECOMP_LU, still should be used if the dimensionality of the space is much smaller than the number of data points. In such a case the overall computing time will be dominated by cv::calcCovarMatrix() anyway. So it may be wise to spend a little bit more time on computing inverse covariance matrix more accurately (much more accurately, if the set of points is concentrated in a subspace of a smaller dimensionality). Thus, DECOMP_EIG is usually the best choice for this task.
23 There is no general function in the library to compute this square root, but because the matrix Σ–1 is very well behaved (as matrices go), the square root can be computed by first diagonalizing it, then taking the square root of the eigenvalues individually, and then rotating back to the original coordinate frame using the same eigenvectors you used to diagonalize it in the first place. (Not surprisingly this technique is called “The Method of Diagonalization”).
24 The problem is that the odds of finding an extremely strange lizard that looks like a dinosaur are still much better than the odds of finding a relatively normal dinosaur. This is because actual dinosaurs are much rarer than lizards.
25 You will notice that ![]() has conspicuously disappeared. This is because not only do we have no prior reason to believe that any value of
has conspicuously disappeared. This is because not only do we have no prior reason to believe that any value of ![]() would be more likely than any other (in the absence of a cluster assignment), but also it would not even matter if that were true, because that factor would be common to all of the things we were comparing.
would be more likely than any other (in the absence of a cluster assignment), but also it would not even matter if that were true, because that factor would be common to all of the things we were comparing.
